Difference between revisions of "Cell Devel Nodes"
| (7 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Infobox Computer | {{Infobox Computer | ||
|image=[[Image:300px-Cell_Broadband_Engine_Processor.jpg|center|300px|thumb]] | |image=[[Image:300px-Cell_Broadband_Engine_Processor.jpg|center|300px|thumb]] | ||
| − | |name=Cell Development | + | |name=Cell Development |
|installed=June 2010 | |installed=June 2010 | ||
|operatingsystem= Linux | |operatingsystem= Linux | ||
| Line 12: | Line 12: | ||
}} | }} | ||
| − | The Cell Development | + | ''' NOTE: THIS SYSTEM IS NO LONGER AVAILABLE ''' |
| − | + | ||
| + | The Cell Development nodes are IBM QS22's each with two 3.2GHz "IBM PowerXCell 8i CPU's, where each CPU has 1 Power Processing Unit (PPU) and 8 Synergistic Processing Units (SPU), and | ||
32GB of RAM per node. | 32GB of RAM per node. | ||
===Login=== | ===Login=== | ||
| − | First login via ssh with your scinet account at <tt>login.scinet.utoronto.ca</tt>, and from there you can proceed to <tt> | + | First login via ssh with your scinet account at '''<tt>login.scinet.utoronto.ca</tt>''', and from there you can proceed to '''<tt>arc01</tt>''' which |
is currently the gateway machine. | is currently the gateway machine. | ||
| Line 25: | Line 26: | ||
You can log into any of 12 nodes '''<tt>blade[03-14]</tt>''' directly to compile/test/run Cell | You can log into any of 12 nodes '''<tt>blade[03-14]</tt>''' directly to compile/test/run Cell | ||
specific or OpenCL codes. | specific or OpenCL codes. | ||
| − | |||
| − | |||
| − | |||
| − | |||
==Programming Frameworks== | ==Programming Frameworks== | ||
Latest revision as of 00:18, 18 September 2013
| Cell Development | |
|---|---|
| Installed | June 2010 |
| Operating System | Linux |
| Interconnect | Infiniband |
| Ram/Node | 32 Gb |
| Cores/Node | 2 PPU + 16 SPU |
| Login/Devel Node | cell-srv01 (from login.scinet) |
| Vendor Compilers | ppu-gcc, spu-gcc |
NOTE: THIS SYSTEM IS NO LONGER AVAILABLE
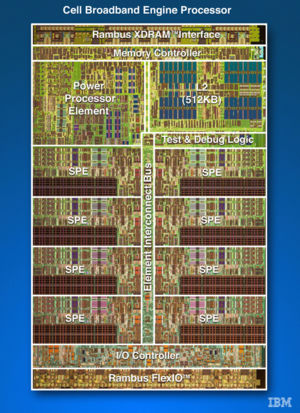
The Cell Development nodes are IBM QS22's each with two 3.2GHz "IBM PowerXCell 8i CPU's, where each CPU has 1 Power Processing Unit (PPU) and 8 Synergistic Processing Units (SPU), and 32GB of RAM per node.
Login
First login via ssh with your scinet account at login.scinet.utoronto.ca, and from there you can proceed to arc01 which is currently the gateway machine.
Compile/Devel/Compute Nodes
Cell
You can log into any of 12 nodes blade[03-14] directly to compile/test/run Cell specific or OpenCL codes.
Programming Frameworks
Currently there are two programming frameworks to use. The Cell SDK is the most full featured and "supported" but is architecture specific, whereas the OpenCL should allow code portability to other accelerators (like GP-GPU's) however is from IBM alphaworks so should be considered beta.
OpenCL
Examples are in
/usr/share/doc/OpenCL-0.1-ibm
Cell SDK
Demos, examples, and build details available in
/opt/cell/sdk/
Compilers
NOTE: gcc has been replaced, use ppu-gcc for most cases where gcc whould be used
- ppu-gcc -- GNU GCC PPC compiler
- spu-gcc -- GNU GCC SPU compiler
- xlcl -- IBM XL OpenCL compiler
MPI
Still a work in progress.
Documentation
- Cell SDK
- Programming:CBE Programmers Guide,CBE Handbook,CBE Programming Tutorial,CBE PXCell Handbook,Performance Tools Reference,IDE Users Guide
- Libraries:3D FFT,libFFT,ALF,BLAS,SIMD Math API,DaCS,LAPACK,Monte Carlo,SDK Library Examples,SPE Runtime Management, SPE Runtime Migration,SPE Runtime Library Ext.
- Architecure: CBEA,CBEA Public Registers,PPC Book 1,PPC Book 2,PPC Book 3,SPU ISA
- Standards:CBE Linux ABI,Language Extentsions,SIMD Library Specification,SPU ABI Specification,SPU Assembly Language Specification
- Installation: Installation Guide
- Cell OpenCL
Further Info
IBM Cell Broadband engine tutorials http://publib.boulder.ibm.com/infocenter/ieduasst/stgv1r0/index.jsp
User Codes
Please discuss put any relevant information/problems/best practices you have encountered when using/developing for CELL and/or OpenCL