Difference between revisions of "HPSS"
m |
m |
||
| (456 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
| + | {| style="border-spacing: 8px; width:100%" | ||
| + | | valign="top" style="cellpadding:1em; padding:1em; border:2px solid; background-color:#f6f674; border-radius:5px"| | ||
| + | '''WARNING: SciNet is in the process of replacing this wiki with a new documentation site. For current information, please go to [https://docs.scinet.utoronto.ca https://docs.scinet.utoronto.ca]''' | ||
| + | |} | ||
| + | |||
| + | {|align=right | ||
| + | |align=center|'''Topology Overview''' | ||
| + | |align=center|'''Submission Queue''' | ||
| + | |- | ||
| + | |[[Image:HPSS-overview.png|right|x200px]] | ||
| + | |[[Image:HPSS-queue2.png|right|x200px]] | ||
| + | |- | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |align=center|'''Servers Rack''' | ||
| + | |align=center|'''TS3500 Library''' | ||
| + | |- | ||
| + | |[[Image:HPSS-servers.png|right|x250px]] | ||
| + | |[[Image:HPSS-TS3500.png|right|x250px]] | ||
| + | |} | ||
| + | |||
= '''High Performance Storage System''' = | = '''High Performance Storage System''' = | ||
| − | ( | + | The High Performance Storage System ([http://www.hpss-collaboration.org/index.shtml HPSS] [http://en.wikipedia.org/wiki/High_Performance_Storage_System wikipedia]) is a tape-backed hierarchical storage system that provides a significant portion of the allocated storage space at SciNet. It is a repository for archiving data that is not being actively used. Data can be returned to the active GPFS filesystem when it is needed. |
| − | + | Since this system is intended for large data storage, it is accessible only to groups who have been awarded storage space at SciNet beyond 5TB in the yearly RAC resource allocation round. However, upon request, any user may be awarded access to HPSS, up to 2TB per group, so that you may get familiar with the system (just email support@scinet.utoronto.ca) | |
Access and transfer of data into and out of HPSS is done under the control of the user, whose interaction is expected to be scripted and submitted as a batch job, using one or more of the following utilities: | Access and transfer of data into and out of HPSS is done under the control of the user, whose interaction is expected to be scripted and submitted as a batch job, using one or more of the following utilities: | ||
| Line 9: | Line 31: | ||
* [http://www.mgleicher.us/GEL/htar HTAR] is a utility that creates tar formatted archives directly into HPSS. It also creates a separate index file (.idx) that can be accessed and browsed quickly. | * [http://www.mgleicher.us/GEL/htar HTAR] is a utility that creates tar formatted archives directly into HPSS. It also creates a separate index file (.idx) that can be accessed and browsed quickly. | ||
* [https://support.scinet.utoronto.ca/wiki/index.php/ISH ISH] is a TUI utility that can perform an inventory of the files and directories in your tarballs. | * [https://support.scinet.utoronto.ca/wiki/index.php/ISH ISH] is a TUI utility that can perform an inventory of the files and directories in your tarballs. | ||
| + | |||
| + | We're currently running HPSS v 7.3.3 patch 6, and HSI/HTAR version 4.0.1.2 | ||
== '''Why should I use and trust HPSS?''' == | == '''Why should I use and trust HPSS?''' == | ||
| − | * | + | * HPSS is a 25 year-old collaboration between IBM and the DoE labs in the US, and is used by about 45 facilities in the [http://www.top500.org “Top 500”] HPC list (plus some black-sites). |
| − | * | + | * Over 2.5 ExaBytes of combined storage world-wide. |
| − | * | + | * The top 3 sites in the World report (fall 2017) having 360PB, 220PB and 125PB in production (ECMWF, UKMO and BNL) |
| + | * Environment Canada also adopted HPSS in 2017 to store Nav Canada data as well as to serve as their own archive. Currently has 2 X 100PB capacity installed. | ||
| + | * The SciNet HPSS system has been providing nearline capacity for important research data in Canada since early 2011, already at 10PB levels in 2018 | ||
| + | * Very reliable, data redundancy and data insurance built-in (dual copies of everything are kept on tapes at SciNet) | ||
| + | * Data on cache and tapes can be geo-distributed for further resilience and HA. | ||
| + | * Highly scalable; current performance at SciNet - after a modest upgrade in 2017 - Ingest: ~150 TB/day, Recall: ~45 TB/day (aggregated). | ||
* HSI/HTAR clients also very reliable and used on several HPSS sites. ISH was written at SciNet. | * HSI/HTAR clients also very reliable and used on several HPSS sites. ISH was written at SciNet. | ||
| − | * [[Media: | + | * [[Media:HPSS_rationale_SNUG.pdf|HPSS fits well with the Storage Capacity Expansion Plan at SciNet]] (pdf presentation) |
== '''Guidelines''' == | == '''Guidelines''' == | ||
| − | * Expanded storage capacity is provided on tape -- a media that is not suited for storing small files. Files smaller than ~ | + | * Expanded storage capacity is provided on tape -- a media that is not suited for storing small files. Files smaller than ~200MB should be grouped into tarballs with tar or htar. |
| − | * | + | * Optimal performance for aggregated transfers and allocation on tapes is obtained with [[Why not tarballs too large |<font color=red>tarballs of size 500GB or less</font>]], whether ingested by htar or hsi ([[Why not tarballs too large | <font color=red>WHY?</font>]]) |
| − | * Make sure to check the application's exit code and | + | * We strongly urge that you use the sample scripts we are providing as the basis for your job submissions. |
| − | + | * Make sure to check the application's exit code and returned logs for errors after any data transfer or tarball creation process | |
| + | |||
| + | == '''New to the System?''' == | ||
| + | The first step is to email scinet support and request an HPSS account (or else you will get "Error - authentication/initialization failed" and 71 exit codes). | ||
| + | |||
| + | THIS set of instructions on the wiki is the best and most compressed "manual" we have. It may seem a bit overwhelming at first, because of all the job script templates we make available below (they are here so you don't have to think | ||
| + | too much, just copy and paste), but if you approach the index at the top as a "case switch" mechanism for what you intend to do, everything falls in place. | ||
| + | |||
| + | Try this sequence: | ||
| + | |||
| + | 1) [https://wiki.scinet.utoronto.ca/wiki/index.php/HPSS#Access_Through_an_Interactive_HSI_session take a look around HPSS using an interactive HSI session] | ||
| + | |||
| + | (most linux shell commands have an equivalent in HPSS) | ||
| + | |||
| + | 2) [https://support.scinet.utoronto.ca/wiki/index.php/HPSS#Sample_tarball_create archive a small test directory using HTAR] | ||
| + | |||
| + | 2a) use step 1) to see what happened | ||
| + | |||
| + | 3) [https://support.scinet.utoronto.ca/wiki/index.php/HPSS#Sample_data_offload archive a file using hsi] | ||
| + | |||
| + | 3a) use step 1) to see what happened | ||
| + | |||
| + | 4) [https://support.scinet.utoronto.ca/wiki/index.php/HPSS#Sample_transferring_directories archive a small test directory using HSI] | ||
| + | |||
| + | 4a) use step 1) to see what happened | ||
| + | |||
| + | 5) now try the other cases and so on. In a couple of hours you'll be in pretty good shape. | ||
| + | |||
| + | == '''Bridge between BGQ and HPSS''' == | ||
| + | |||
| + | At this time BGQ users will have to migrate data to Niagara scratch prior to transferring it to HPSS. We are looking for ways to improve this workflow. | ||
== '''Access Through the Queue System''' == | == '''Access Through the Queue System''' == | ||
| − | All access to the archive system is done through the [ | + | All access to the archive system is done through the [https://docs.computecanada.ca/wiki/Niagara_Quickstart#Submitting_jobs NIA queue system]. |
| + | |||
| + | * Job submissions should be done to the 'archivelong' queue or the 'archiveshort' | ||
| + | * Short jobs are limited to 1H walltime by default. Long jobs (> 1H) are limited to 72H walltime. | ||
| + | * Users are limited to only 2 long jobs and 2 short jobs at the same time, and 10 jobs total on the each queue. | ||
| + | * There can only be 5 long jobs running at any given time overall. Remaining submissions will be placed on hold for the time being. So far we have not seen a need for overall limit on short jobs. | ||
| + | |||
| + | The status of pending jobs can be monitored with squeue specifying the archive queue: | ||
| + | <pre> | ||
| + | squeue -p archiveshort | ||
| + | OR | ||
| + | squeue -p archivelong | ||
| + | </pre> | ||
| + | |||
| + | == '''Access Through an Interactive HSI session''' == | ||
| + | * You may want to acquire an interactive shell, start an HSI session and navigate the archive naming-space. Keep in mind, you're restricted to 1H. | ||
| + | |||
| + | <pre> | ||
| + | pinto@nia-login07:~$ salloc -p archiveshort -t 1:00:00 | ||
| + | salloc: Granted job allocation 50918 | ||
| + | salloc: Waiting for resource configuration | ||
| + | salloc: Nodes hpss-archive02-ib are ready for job | ||
| + | hpss-archive02-ib:~$ | ||
| + | |||
| + | hpss-archive02-ib:~$ hsi (DON'T FORGET TO START HSI) | ||
| + | ****************************************************************** | ||
| + | * Welcome to HPSS@SciNet - High Perfomance Storage System * | ||
| + | * * | ||
| + | * INFO: THIS IS THE NEW 7.5.1 HPSS SYSTEM! * | ||
| + | * * | ||
| + | * Contact Information: support@scinet.utoronto.ca * | ||
| + | * NOTE: do not transfer SMALL FILES with HSI. Use HTAR instead * | ||
| + | * CHECK THE INTEGRITY OF YOUR TARBALLS * | ||
| + | ****************************************************************** | ||
| + | [HSI]/archive/s/scinet/pinto-> ls | ||
| + | |||
| + | [HSI]/archive/s/scinet/pinto-> cd <some directory> | ||
| + | </pre> | ||
| + | |||
=== Scripted File Transfers === | === Scripted File Transfers === | ||
| − | File transfers in and out of the HPSS should be scripted into jobs and submitted to the '' | + | File transfers in and out of the HPSS should be scripted into jobs and submitted to the ''archivelong'' queue or the ''archiveshort'' . See generic example below: |
| − | < | + | |
| − | #!/bin/ | + | <source lang="bash"> |
| − | # | + | #!/bin/bash -l |
| − | # | + | #SBATCH -t 72:00:00 |
| − | # | + | #SBATCH -p archivelong |
| − | # | + | #SBATCH -N 1 |
| + | #SBATCH -J htar_create_tarball_in_hpss | ||
| + | #SBATCH --mail-type=ALL | ||
| + | |||
| + | echo "Creating a htar of finished-job1/ directory tree into HPSS" | ||
| + | |||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | # Note that your initial directory in HPSS will be $ARCHIVE | ||
| + | |||
| + | DEST=$ARCHIVE/finished-job1.tar | ||
| + | |||
| + | # htar WILL overwrite an existing file with the same name so check beforehand. | ||
| + | |||
| + | hsi ls $DEST &> /dev/null | ||
| + | status=$? | ||
| + | |||
| + | if [ $status == 0 ]; then | ||
| + | echo 'File $DEST already exists. Nothing has been done' | ||
| + | exit 1 | ||
| + | fi | ||
| − | / | + | cd $SCRATCH/workarea/ |
| − | + | htar -Humask=0137 -cpf $ARCHIVE/finished-job1.tar finished-job1/ | |
| − | |||
status=$? | status=$? | ||
| − | if [ ! $status == 0 ];then | + | |
| − | echo ' | + | trap - TERM INT |
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HTAR returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
fi | fi | ||
| − | exit | + | </source> |
| + | '''Note:''' Always trap the execution of your jobs for abnormal terminations, and be sure to return the exit code | ||
| + | |||
| + | === Job Dependencies === | ||
| + | |||
| + | Typically data will be recalled to /scratch when it is needed for analysis. Job dependencies can be constructed so that analysis jobs wait in the queue for data recalls before starting. The qsub flag is | ||
| + | <pre> | ||
| + | --dependency=<type:JOBID> | ||
</pre> | </pre> | ||
| + | where JOBID is the job number of the archive recalling job that must finish successfully before the analysis job can start. | ||
| − | + | Here is a short cut for generating the dependency (lookup [https://support.scinet.utoronto.ca/wiki/index.php/HPSS#Sample_data_recall data-recall.sh samples]): | |
<pre> | <pre> | ||
| − | + | hpss-archive02-ib:~$ sbatch $(sbatch data-recall.sh | awk {print "--dependency=afterany:"$1}') job-to-work-on-recalled-data.sh | |
</pre> | </pre> | ||
| − | == | + | == '''HTAR''' == |
| + | ''' Please aggregate small files (<~200MB) into tarballs or htar files. ''' | ||
| − | + | ''' [[Why not tarballs too large |<font color=red>Keep your tarballs to size 500GB or less</font>]], whether ingested by htar or hsi ([[Why not tarballs too large | <font color=red>WHY?</font>]])''' | |
| + | |||
| + | HTAR is a utility that is used for aggregating a set of files and directories, by using a sophisticated multithreaded buffering scheme to write files directly from GPFS into HPSS, creating an archive file that conforms to the POSIX TAR specification, thereby achieving a high rate of performance. HTAR does not do gzip compression, however it already has a built-in checksum algorithm. | ||
| + | |||
| + | '''Caution''' | ||
| + | * Files larger than 68 GB cannot be stored in an HTAR archive. If you attempt to start a transfer with any files larger than 68GB the whole HTAR session will fail, and you'll get a notification listing all those files, so that you can transfer them with HSI. | ||
| + | * Files with pathnames too long will be skipped (greater than 100 characters), so as to conform with TAR protocol [[(POSIX 1003.1 USTAR)]] -- Note that the HTAR will erroneously indicate success, however will produce exit code 70. For now, you can check for this type of error by "grep Warning my.output" after the job has completed. | ||
| + | * Unlike with cput/cget in HSI, "prompt before overwrite", this is not the default with (h)tar. Be careful not to unintentionally overwrite a previous htar destination file in HPSS. There could be a similar situation when extracting material back into GPFS and overwriting the originals. Be sure to double-check the logic in your scripts. | ||
| + | * Check the HTAR exit code and log file before removing any files from the GPFS active filesystems. | ||
| + | |||
| + | |||
| + | === HTAR Usage === | ||
| + | * To write the ''file1'' and ''file2'' files to a new archive called ''files.tar'' in the default HPSS home directory, and preserve mask attributes (-p), enter: | ||
<pre> | <pre> | ||
| − | - | + | htar -cpf files.tar file1 file2 |
| + | OR | ||
| + | htar -cpf $ARCHIVE/files.tar file1 file2 | ||
</pre> | </pre> | ||
| − | |||
| − | + | * To write a ''subdirA'' to a new archive called ''subdirA.tar'' in the default HPSS home directory, enter: | |
<pre> | <pre> | ||
| − | + | htar -cpf subdirA.tar subdirA | |
</pre> | </pre> | ||
| − | + | * To extract all files from the archive file called ''proj1.tar'' in HPSS into the ''project1/src'' directory in GPFS, and use the time of extraction as the modification time, enter: | |
| + | <pre> | ||
| + | cd project1/src | ||
| + | htar -xpmf proj1.tar | ||
| + | </pre> | ||
| − | HSI | + | * To display the names of the files in the ''out.tar'' archive file within the HPSS home directory, enter (the out.tar.idx file will be queried): |
| + | <pre> | ||
| + | htar -vtf out.tar | ||
| + | </pre> | ||
| + | |||
| + | * To ensure that both the htar and the .idx files have read permissions to other members in your group use the umask option | ||
| + | <pre> | ||
| + | htar -Humask=0137 .... | ||
| + | </pre> | ||
| + | |||
| + | For more details please check the '''[http://www.mgleicher.us/GEL/htar/ HTAR - Introduction]''' or the '''[http://www.mgleicher.us/GEL/htar/htar_man_page.html HTAR Man Page]''' online | ||
| + | |||
| + | |||
| + | ==== Sample tarball create ==== | ||
| + | <source lang="bash"> | ||
| + | #!/bin/bash -l | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J htar_create_tarball_in_hpss | ||
| + | #SBATCH --mail-type=ALL | ||
| + | |||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | # Note that your initial directory in HPSS will be /archive/$(id -gn)/$(whoami)/ | ||
| + | |||
| + | DEST=$ARCHIVE/finished-job1.tar | ||
| + | |||
| + | # htar WILL overwrite an existing file with the same name so check beforehand. | ||
| + | |||
| + | hsi ls $DEST &> /dev/null | ||
| + | status=$? | ||
| + | |||
| + | if [ $status == 0 ]; then | ||
| + | echo 'File $DEST already exists. Nothing has been done' | ||
| + | exit 1 | ||
| + | fi | ||
| + | |||
| + | cd $SCRATCH/workarea/ | ||
| + | htar -Humask=0137 -cpf $DEST finished-job1/ | ||
| + | status=$? | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HTAR returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | '''Note:''' If you attempt to start a transfer with any files larger than 68GB the whole HTAR session will fail, and you'll get a notification listing all those files, so that you can transfer them with HSI. | ||
| + | <pre> | ||
| + | ---------------------------------------- | ||
| + | INFO: File too large for htar to handle: finished-job1/file1 (86567185745 bytes) | ||
| + | INFO: File too large for htar to handle: finished-job1/file2 (71857244579 bytes) | ||
| + | ERROR: 2 oversize member files found - please correct and retry | ||
| + | ERROR: [FATAL] error(s) generating filename list | ||
| + | HTAR: HTAR FAILED | ||
| + | ###WARNING htar returned non-zero exit status | ||
| + | </pre> | ||
| + | |||
| + | ==== Sample tarball list ==== | ||
| + | <source lang="bash"> | ||
| + | #!/bin/bash -l | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J htar_list_tarball_in_hpss | ||
| + | #SBATCH --mail-type=ALL | ||
| + | |||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | # Note that your initial directory in HPSS will be $ARCHIVE | ||
| + | |||
| + | DEST=$ARCHIVE/finished-job1.tar | ||
| + | |||
| + | htar -tvf $DEST | ||
| + | status=$? | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HTAR returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | ==== Sample tarball extract ==== | ||
| + | <source lang="bash"> | ||
| + | #!/bin/bash | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J htar_extract_tarball_from_hpss | ||
| + | #SBATCH --mail-type=ALL | ||
| + | |||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | # Note that your initial directory in HPSS will be $ARCHIVE | ||
| + | |||
| + | cd $SCRATCH/recalled-from-hpss | ||
| + | htar -xpmf $ARCHIVE/finished-job1.tar | ||
| + | status=$? | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HTAR returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | == '''HSI''' == | ||
| + | |||
| + | HSI may be the primary client with which some users will interact with HPSS. It provides an ftp-like interface for archiving and retrieving tarballs or [https://support.scinet.utoronto.ca/wiki/index.php/HPSS#Sample_transferring_directories directory trees]. In addition it provides a number of shell-like commands that are useful for examining and manipulating the contents in HPSS. The most commonly used commands will be: | ||
{|border="1" cellpadding="10" cellspacing="0" | {|border="1" cellpadding="10" cellspacing="0" | ||
|- | |- | ||
| cput | | cput | ||
| − | | Conditionally | + | | Conditionally saves or replaces a HPSSpath file to GPFSpath if the GPFS version is new or has been updated |
| + | cput [options] GPFSpath [: HPSSpath] | ||
|- | |- | ||
| cget | | cget | ||
| − | | Conditionally retrieves a copy of a file from HPSS to | + | | Conditionally retrieves a copy of a file from HPSS to GPFS only if a GPFS version does not already exist. |
| + | cget [options] [GPFSpath :] HPSSpath | ||
|- | |- | ||
| cd,mkdir,ls,rm,mv | | cd,mkdir,ls,rm,mv | ||
| Line 77: | Line 347: | ||
|- | |- | ||
| lcd,lls | | lcd,lls | ||
| − | | ''Local'' commands | + | | ''Local'' commands to GPFS |
|} | |} | ||
| − | * | + | |
| + | *There are 3 distinctions about HSI that you should keep in mind, and that can generate a bit of confusion when you're first learning how to use it: | ||
| + | ** HSI doesn't currently support renaming directories paths during transfers on-the-fly, therefore the syntax for cput/cget may not work as one would expect in some scenarios, requiring some workarounds. | ||
| + | ** HSI has an operator ":" which separates the GPFSpath and HPSSpath, and must be surrounded by whitespace (one or more space characters) | ||
| + | ** The order for referring to files in HSI syntax is different from FTP. In HSI the general format is always the same, GPFS first, HPSS second, cput or cget: | ||
<pre> | <pre> | ||
| − | + | GPFSfile : HPSSfile | |
</pre> | </pre> | ||
| − | + | For example, when using HSI to store the tarball file from GPFS into HPSS, then recall it to GPFS, the following commands could be used: | |
<pre> | <pre> | ||
| − | + | cput tarball-in-GPFS : tarball-in-HPSS | |
| − | + | cget tarball-recalled : tarball-in-HPSS | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</pre> | </pre> | ||
| − | + | unlike with FTP, where the following syntax would be used: | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<pre> | <pre> | ||
| − | + | put tarball-in-GPFS tarball-in-HPSS | |
| − | + | get tarball-in-HPSS tarball-recalled | |
| − | |||
</pre> | </pre> | ||
| − | + | * Simple commands can be executed on a single line. | |
| − | |||
<pre> | <pre> | ||
| − | + | hsi "mkdir LargeFilesDir; cd LargeFilesDir; cput tarball-in-GPFS : tarball-in-HPSS" | |
</pre> | </pre> | ||
| − | + | * More complex sequences can be performed using an except such as this: | |
<pre> | <pre> | ||
| − | / | + | hsi <<EOF |
| + | mkdir LargeFilesDir | ||
| + | cd LargeFilesDir | ||
| + | cput tarball-in-GPFS : tarball-in-HPSS | ||
| + | lcd $SCRATCH/LargeFilesDir2/ | ||
| + | cput -Ruph * | ||
| + | end | ||
| + | EOF | ||
</pre> | </pre> | ||
| − | + | * The commands below are equivalent, but we recommend that you always use full path, and organize the contents of HPSS, where the default HSI directory placement is $ARCHIVE: | |
<pre> | <pre> | ||
| − | + | hsi cput tarball | |
| + | hsi cput tarball : tarball | ||
| + | hsi cput $SCRATCH/tarball : $ARCHIVE/tarball | ||
</pre> | </pre> | ||
| − | + | * There are no known issues renaming files on-the-fly: | |
<pre> | <pre> | ||
| − | + | hsi cput $SCRATCH/tarball1 : $ARCHIVE/tarball2 | |
| + | hsi cget $SCRATCH/tarball3 : $ARCHIVE/tarball2 | ||
</pre> | </pre> | ||
| − | + | * However the syntax forms such as the ones below will fail, since they rename the directory paths. | |
<pre> | <pre> | ||
| − | + | hsi cput -Ruph $SCRATCH/LargeFilesDir : $ARCHIVE/LargeFilesDir (FAILS) | |
| + | OR | ||
| + | hsi cget -Ruph $SCRATCH/LargeFilesDir : $ARCHIVE/LargeFilesDir2 (FAILS) | ||
| + | OR | ||
| + | hsi cput -Ruph $SCRATCH/LargeFilesDir/* : $ARCHIVE/LargeFilesDir2 (FAILS) | ||
| + | OR | ||
| + | hsi cget -Ruph $SCRATCH/LargeFilesDir : $ARCHIVE/LargeFilesDir (FAILS) | ||
</pre> | </pre> | ||
| − | |||
| − | + | One workaround is the following 2-steps process, where you do a "lcd " in GPFS first, and recursively transfer the whole directory (-R), keeping the same name. You may use '-u' option to resume a previously disrupted session, and the '-p' to preserve timestamp, and '-h' to keep the links. | |
<pre> | <pre> | ||
| − | + | hsi <<EOF | |
| + | lcd $SCRATCH | ||
| + | cget -Ruph LargeFilesDir | ||
| + | end | ||
| + | EOF | ||
</pre> | </pre> | ||
| − | + | Another workaround is do a "lcd" into the GPFSpath first and a "cd" in the HPSSpath, but transfer the files individually with the '*' wild character. This option lets you change the directory name: | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<pre> | <pre> | ||
| + | hsi <<EOF | ||
| + | lcd $SCRATCH/LargeFilesDir | ||
| + | mkdir $ARCHIVE/LargeFilesDir2 | ||
| + | cd $ARCHIVE/LargeFilesDir2 | ||
| + | cput -Ruph * | ||
| + | end | ||
| + | EOF | ||
</pre> | </pre> | ||
| + | === Documentation === | ||
| + | Complete documentation on HSI is available from the Gleicher Enterprises links below. You may peruse those links and come with alternative syntax forms. You may even be already familiar with HPSS/HSI from other HPC facilities, that may or not have procedures similar to ours. HSI doesn't always work as expected when you go outside of our recommended syntax, so '''we strongly urge that you use the sample scripts we are providing as the basis''' for your job submissions | ||
| + | * [http://www.mgleicher.us/hsi/hsi_reference_manual_2/introduction.html HSI Introduction] | ||
| + | * [http://www.mgleicher.us/hsi/hsi_man_page.html man hsi] | ||
| + | * [http://support.scinet.utoronto.ca/wiki/index.php/HSI_help hsi help] | ||
| + | * [http://www.mgleicher.us/hsi/hsi-exit-codes.html exit codes] | ||
| + | '''Note:''' HSI returns the highest-numbered exit code, in case of multiple operations in the same hsi session. You may use '/scinet/niagara/bin/exit2msg $status' to translate those codes into intelligible messages | ||
| − | + | === Typical Usage Scripts=== | |
| − | + | The most common interactions will be ''putting'' data into HPSS, examining the contents (ls,ish), and ''getting'' data back onto GPFS for inspection or analysis. | |
| − | === | + | ==== Sample '''data offload''' ==== |
| − | + | <source lang="bash"> | |
| − | |||
| − | |||
| − | < | ||
#!/bin/bash | #!/bin/bash | ||
| − | |||
# This script is named: data-offload.sh | # This script is named: data-offload.sh | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J offload | ||
| + | #SBATCH --mail-type=ALL | ||
| − | + | trap "echo 'Job script not completed';exit 129" TERM INT | |
| − | + | # individual tarballs already exist | |
| − | |||
| − | # | ||
| − | + | /usr/local/bin/hsi -v <<EOF1 | |
| + | mkdir put-away | ||
| + | cd put-away | ||
| + | cput $SCRATCH/workarea/finished-job1.tar.gz : finished-job1.tar.gz | ||
| + | end | ||
| + | EOF1 | ||
| + | status=$? | ||
| + | if [ ! $status == 0 ];then | ||
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| − | + | /usr/local/bin/hsi -v <<EOF2 | |
| − | |||
| − | /usr/local/bin/hsi -v << | ||
mkdir put-away | mkdir put-away | ||
cd put-away | cd put-away | ||
| − | cput | + | cput $SCRATCH/workarea/finished-job2.tar.gz : finished-job2.tar.gz |
| − | + | end | |
| − | + | EOF2 | |
status=$? | status=$? | ||
if [ ! $status == 0 ];then | if [ ! $status == 0 ];then | ||
| − | echo '! | + | echo 'HSI returned non-zero code.' |
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | |||
| + | trap - TERM INT | ||
| + | </source> | ||
| + | |||
| + | |||
| + | '''Note:''' as in the above example, we recommend that you capture the (highest-numbered) exit code for each hsi session independently. And remember, you may improve your exit code verbosity by adding the excerpt below to your scripts: | ||
| + | <source lang="bash"> | ||
| + | if [ ! $status == 0 ];then | ||
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
fi | fi | ||
| − | + | </source> | |
| − | </ | ||
| − | + | ==== Sample '''data list''' ==== | |
| − | A | + | A very trivial way to list the contents of HPSS would be to just submit the HSI 'ls' command. |
| − | < | + | <source lang="bash"> |
#!/bin/bash | #!/bin/bash | ||
| + | # This script is named: data-list.sh | ||
| + | #SBATCH -t 1:00:00 | ||
| + | #SBATCH -p archiveshort | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J hpss_ls | ||
| + | #SBATCH --mail-type=ALL | ||
| + | /usr/local/bin/hsi -v <<EOF | ||
| + | cd put-away | ||
| + | ls -R | ||
| + | end | ||
| + | EOF | ||
| + | </source> | ||
| + | ''Warning: if you have a lot of files, the ls command will take a long time to complete. For instance, about 400,000 files can be listed in about an hour. Adjust the walltime accordingly, and be on the safe side.'' | ||
| + | |||
| + | However, we provide a much more useful and convenient way to explore the contents of HPSS with the inventory shell [[ISH]]. This example creates an index of all the files in a user's portion of the namespace. The list is placed in the directory /home/$(whoami)/.ish_register that can be inspected from the login nodes. | ||
| + | <source lang="bash"> | ||
| + | #!/bin/bash | ||
# This script is named: data-list.sh | # This script is named: data-list.sh | ||
| + | #SBATCH -t 1:00:00 | ||
| + | #SBATCH -p archiveshort | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J hpss_index | ||
| + | #SBATCH --mail-type=ALL | ||
| − | + | INDEX_DIR=$HOME/.ish_register | |
| − | + | if ! [ -e "$INDEX_DIR" ]; then | |
| − | + | mkdir -p $INDEX_DIR | |
| − | |||
| − | |||
| − | |||
| − | INDEX_DIR= | ||
| − | if | ||
| − | mkdir $INDEX_DIR | ||
fi | fi | ||
| − | export ISHREGISTER=$ | + | export ISHREGISTER="$INDEX_DIR" |
| − | ish hindex | + | /scinet/niagara/bin/ish hindex |
| − | </ | + | </source> |
| + | ''Note: the above warning on collecting the listing for many files applies here too.'' | ||
This index can be browsed or searched with ISH on the development nodes. | This index can be browsed or searched with ISH on the development nodes. | ||
| − | < | + | <source lang="bash"> |
| − | + | hpss-archive02-ib:~$ /scinet/niagara/bin/ish ~/.ish_register/hpss.igz | |
| − | [ish] | + | [ish]hpss.igz> help |
| − | </ | + | </source> |
ISH is a powerful tool that is also useful for creating and browsing indices of tar and htar archives, so please look at the [[ISH|documentation]] or built in help. | ISH is a powerful tool that is also useful for creating and browsing indices of tar and htar archives, so please look at the [[ISH|documentation]] or built in help. | ||
| − | + | ==== Sample '''data recall''' ==== | |
| − | - | + | <source lang="bash"> |
| + | #!/bin/bash | ||
| + | # This script is named: data-recall.sh | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J recall_files | ||
| + | #SBATCH --mail-type=ALL | ||
| + | |||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | |||
| + | mkdir -p $SCRATCH/recalled-from-hpss | ||
| + | |||
| + | # individual tarballs previously organized in HPSS inside the put-away-on-2010/ folder | ||
| + | hsi -v << EOF | ||
| + | cget $SCRATCH/recalled-from-hpss/Jan-2010-jobs.tar.gz : $ARCHIVE/put-away-on-2010/Jan-2010-jobs.tar.gz | ||
| + | cget $SCRATCH/recalled-from-hpss/Feb-2010-jobs.tar.gz : $ARCHIVE/put-away-on-2010/Feb-2010-jobs.tar.gz | ||
| + | end | ||
| + | EOF | ||
| + | status=$? | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagar/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | |||
| + | We should emphasize that a single ''cget'' of multiple files (rather than several separate gets) allows HSI to do optimization, as in the following example: | ||
| + | <source lang="bash"> | ||
| + | #!/bin/bash | ||
| + | # This script is named: data-recall.sh | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J recall_files_optimized | ||
| + | #SBATCH --mail-type=AL | ||
| + | |||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | mkdir -p $SCRATCH/recalled-from-hpss | ||
| + | |||
| + | # individual tarballs previously organized in HPSS inside the put-away-on-2010/ folder | ||
| + | hsi -v << EOF | ||
| + | lcd $SCRATCH/recalled-from-hpss/ | ||
| + | cd $ARCHIVE/put-away-on-2010/ | ||
| + | cget Jan-2010-jobs.tar.gz Feb-2010-jobs.tar.gz | ||
| + | end | ||
| + | EOF | ||
| + | status=$? | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | ==== Sample '''transferring directories''' ==== | ||
| + | Remember, it's not possible to rename directories on-the-fly: | ||
<pre> | <pre> | ||
| + | hsi cget -Ruph $SCRATCH/LargeFiles-recalled : $ARCHIVE/LargeFiles (FAILS) | ||
| + | </pre> | ||
| + | |||
| + | One workaround is transfer the whole directory (and sub-directories) recursively: | ||
| + | <source lang="bash"> | ||
#!/bin/bash | #!/bin/bash | ||
| + | # This script is named: data-recall.sh | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J recall_directories | ||
| + | #SBATCH --mail-type=ALL | ||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | |||
| + | mkdir -p $SCRATCH/recalled | ||
| + | |||
| + | hsi -v << EOF | ||
| + | lcd $SCRATCH/recalled | ||
| + | cd $ARCHIVE/ | ||
| + | cget -Ruph LargeFiles | ||
| + | end | ||
| + | EOF | ||
| + | status=$? | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | |||
| + | Another workaround is to transfer files and subdirectories individually with the "*" wild character: | ||
| + | <source lang="bash"> | ||
| + | #!/bin/bash | ||
# This script is named: data-recall.sh | # This script is named: data-recall.sh | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J recall_directories | ||
| + | #SBATCH --mail-type=AL | ||
| − | + | trap "echo 'Job script not completed';exit 129" TERM INT | |
| − | |||
| − | |||
| − | |||
| + | mkdir -p $SCRATCH/LargeFiles-recalled | ||
| − | |||
hsi -v << EOF | hsi -v << EOF | ||
| − | + | lcd $SCRATCH/LargeFiles-recalled | |
| − | cget | + | cd $ARCHIVE/LargeFiles |
| + | cget -Ruph * | ||
| + | end | ||
EOF | EOF | ||
status=$? | status=$? | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | |||
| + | |||
| + | * For more details please check the '''[http://www.mgleicher.us/GEL/hsi/ HSI Introduction]''', the '''[http://www.mgleicher.us/GEL/hsi/hsi_man_page.html HSI Man Page]''' or the or the [https://support.scinet.utoronto.ca/wiki/index.php/HSI_help '''hsi help'''] | ||
| + | |||
| + | == '''[[ISH|ISH]]''' == | ||
| + | === [[ISH|Documentation and Usage]] === | ||
| + | |||
| + | == '''File and directory management''' == | ||
| + | === Moving/renaming === | ||
| + | * you may use 'mv' or 'cp' in the same way as the linux version. | ||
| + | <source lang="bash"> | ||
| + | #!/bin/bash | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J deletion_script | ||
| + | #SBATCH --mail-type=ALL | ||
| + | |||
| + | echo "HPSS file and directory management" | ||
| + | |||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | |||
| + | /usr/local/bin/hsi -v <<EOF1 | ||
| + | mkdir $ARCHIVE/2011 | ||
| + | mv $ARCHIVE/oldjobs $ARCHIVE/2011 | ||
| + | cp -r $ARCHIVE/almostfinished/*done $ARCHIVE/2011 | ||
| + | end | ||
| + | EOF1 | ||
| + | status=$? | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | === Deletions === | ||
| + | ==== Recommendations ==== | ||
| + | * Be careful with the use of 'cd' commands to non-existing directories before the 'rm' command. Results may be unpredictable | ||
| + | * Avoid the use of the stand alone wild character '''*'''. If necessary, whenever possible have it bound to common patterns, such as '*.tmp', so to limit unintentional mis-happens | ||
| + | * Avoid using relative paths, even the env variable $ARCHIVE. Better to explicitly expand the full paths in your scripts | ||
| + | * Avoid using recursive/looped deletion instructions on $SCRATCH contents from the archive job scripts. Even on $ARCHIVE contents, it may be better to do it as an independent job submission, after you have verified that the original ingestion into HPSS finished without any issues. | ||
| + | |||
| + | ==== Typical example ==== | ||
| + | <source lang="bash"> | ||
| + | #!/bin/bash | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J deletion_script | ||
| + | #SBATCH --mail-type=ALL | ||
| − | + | echo "Deletion of an outdated directory tree into HPSS" | |
| − | </ | + | trap "echo 'Job script not completed';exit 129" TERM INT |
| + | # Note that the initial directory in HPSS ($ARCHIVE) has the path explicitly expanded | ||
| + | |||
| + | /usr/local/bin/hsi -v <<EOF1 | ||
| + | rm /archive/s/scinet/pinto/*.tmp | ||
| + | rm -R /archive/s/scinet/pinto/obsolete | ||
| + | end | ||
| + | EOF1 | ||
| + | status=$? | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | ==== Deleting with an interactive HSI session ==== | ||
| + | * You may feel more comfortable acquiring an interactive shell, starting an HSI session and proceeding with your deletions that way. Keep in mind, you're restricted to 1H. | ||
| − | + | * After using the ''qsub -q archive -I'' command you'll get a standard shell prompt on an archive execution node (hpss-archive02), as you would on any compute node. However you will need to run '''HSI''' or '''HTAR''' to access resources on HPSS. | |
| − | HSI | ||
| − | HSI | + | * HSI will give you a prompt very similar to a standard shell, where your can navigate around using commands such 'ls', 'cd', 'pwd', etc ... NOTE: not every bash command has an equivalent on HSI - for instance, you can not 'vi' or 'cat'. |
| − | |||
| − | |||
| − | |||
| − | |||
<pre> | <pre> | ||
| − | + | pinto@nia-login07:~$ salloc -p archiveshort -t 1:00:00 | |
| + | salloc: Granted job allocation 50359 | ||
| + | salloc: Waiting for resource configuration | ||
| + | salloc: Nodes hpss-archive02-ib are ready for job | ||
| + | |||
| + | hpss-archive02-ib:~$ hsi | ||
| + | ****************************************************************** | ||
| + | * Welcome to HPSS@SciNet - High Perfomance Storage System * | ||
| + | * * | ||
| + | * INFO: THIS IS THE NEW 7.5.1 HPSS SYSTEM! * | ||
| + | * * | ||
| + | * Contact Information: support@scinet.utoronto.ca * | ||
| + | * NOTE: do not transfer SMALL FILES with HSI. Use HTAR instead * | ||
| + | * CHECK THE INTEGRITY OF YOUR TARBALLS * | ||
| + | ****************************************************************** | ||
| + | |||
| + | [HSI]/archive/s/scinet/pinto-> rm -R junk | ||
| + | |||
</pre> | </pre> | ||
| − | |||
| − | + | == '''HPSS for the 'Watchmaker' ''' == | |
| − | < | + | === Efficient alternative to htar === |
| − | + | <source lang="bash"> | |
| − | + | #!/bin/bash | |
| − | </ | + | #SBATCH -t 72:00:00 |
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J tar_create_tarball_in_hpss_with_hsi_by_piping | ||
| + | #SBATCH --mail-type=ALL | ||
| + | |||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | # Note that your initial directory in HPSS will be $ARCHIVE | ||
| + | |||
| + | # When using a pipeline like this | ||
| + | set -o pipefail | ||
| + | |||
| + | # to put (cput will fail) | ||
| + | tar -c $SCRATCH/mydir | hsi put - : $ARCHIVE/mydir.tar | ||
| + | status=$? | ||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'TAR+HSI+piping returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | |||
| + | # to immediately generate an index | ||
| + | ish hindex $ARCHIVE/mydir.tar | ||
| + | status=$? | ||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'ISH returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | |||
| + | # to get | ||
| + | #cd $SCRATCH | ||
| + | #hsi cget - : $ARCHIVE/mydir.tar | tar -xv | ||
| + | #status=$? | ||
| + | # if [ ! $status == 0 ]; then | ||
| + | # echo 'TAR+HSI+piping returned non-zero code.' | ||
| + | # /scinet/niagara/bin/exit2msg $status | ||
| + | # exit $status | ||
| + | #else | ||
| + | # echo 'TRANSFER SUCCESSFUL' | ||
| + | #fi | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | </source> | ||
| + | '''Notes:''' | ||
| + | * Combining commands in this fashion, besides being HPSS-friendly, should not be that noticeably slower than the recursive put with HSI that stores each file one by one. However, reading the files back from tape in this format will be many times faster. It would also overcome the current 68GB limit on the size of stored files that we have with htar. | ||
| + | * To top things off, we recommend indexing with ish (in the same script) immediately after the tarball creation , while it resides in the HPSS cache. It would be as if htar was used. | ||
| + | * To ensure that an error at any stage of the pipeline shows up in the returned status use: ''set -o pipefail'' (The default is to return the status of the last command in the pipeline and this is not what you want.) | ||
| + | * Optimal performance for aggregated transfers and allocation on tapes is obtained with [[Why not tarballs too large |<font color=red>tarballs of size 500GB or less</font>]], whether ingested by htar or hsi ([[Why not tarballs too large | <font color=red>WHY?</font>]]). Be sure to check the contents of the directory tree with 'du' for the total amount of data before sending them to the tar+HSI piping. | ||
| + | |||
| + | === Multi-threaded gzip'ed compression with pigz === | ||
| + | We compiled multi-threaded implementation of gzip called pigz (http://zlib.net/pigz/). It's now part of the "extras" module. It can also be used on any compute or devel nodes. This makes the execution of the previous version of the script much quicker than if you were to use 'tar -cfz'. In addition, by piggy-backing ISH to the end of the script, it will know what to do with the just created mydir.tar.gz compressed tarball. | ||
| + | |||
| + | <source lang="bash"> | ||
| + | #!/bin/bash | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J tar_create_compressed_tarball_in_hpss_with_hsi_by_piping | ||
| + | #SBATCH --mail-type=ALL | ||
| + | |||
| + | |||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | # Note that your initial directory in HPSS will be $ARCHIVE | ||
| + | |||
| + | # When using a pipeline like this | ||
| + | set -o pipefail | ||
| + | |||
| + | load module extras | ||
| + | |||
| + | # to put (cput will fail) | ||
| + | tar -c $SCRATCH/mydir | pigz | hsi put - : $ARCHIVE/mydir.tar.gz | ||
| + | status=$? | ||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'TAR+PIGZ+HSI+piping returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | === Content Verification === | ||
| + | |||
| + | ==== HTAR CRC checksums ==== | ||
| + | Specifies that HTAR should generate CRC checksums when creating the archive. | ||
| + | |||
| + | <source lang="bash"> | ||
| + | #!/bin/bash | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J htar_create_tarball_in_hpss_with_checksum_verification | ||
| + | #SBATCH --mail-type=ALL | ||
| + | |||
| + | trap "echo 'Job script not completed';exit 129" TERM INT | ||
| + | # Note that your initial directory in HPSS will be $ARCHIVE | ||
| + | |||
| + | cd $SCRATCH/workarea | ||
| + | |||
| + | # to put | ||
| + | htar -Humask=0137 -cpf $ARCHIVE/finished-job1.tar -Hcrc -Hverify=1 finished-job1/ | ||
| + | |||
| + | # to get | ||
| + | #mkdir $SCRATCH/verification | ||
| + | #cd $SCRATCH/verification | ||
| + | #htar -Hcrc -xvpmf $ARCHIVE/finished-job1.tar | ||
| + | |||
| + | status=$? | ||
| + | |||
| + | trap - TERM INT | ||
| + | |||
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HTAR returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| + | |||
| + | ==== Current HSI version - Checksum built-in ==== | ||
| + | |||
| + | MD5 is the standard Hashing Algorithm for the HSI build at SciNet. For hsi ingestions with the '-c on' option you should be able to query the md5 hash with the hsi command 'hashli'. That value is stored as an UDA (User Defined Attribute) for each file (a feature of HPSS starting with 7.4) | ||
| + | |||
| + | [http://www.mgleicher.us/GEL/hsi/hsi/hsi_reference_manual_2/checksum-feature.html More usage details here] | ||
| + | |||
| + | The checksum algorithm is very CPU-intensive. Although the checksum code is compiled with a high level of compiler optimization, transfer rates can be significantly reduced when checksum creation or verification is in effect. The amount of degradation in transfer rates depends on several factors, such as processor speed, network transfer speed, and speed of the local filesystem (GPFS). | ||
| + | |||
| + | <source lang="bash"> | ||
| + | #!/bin/bash | ||
| + | #SBATCH -t 72:00:00 | ||
| + | #SBATCH -p archivelong | ||
| + | #SBATCH -N 1 | ||
| + | #SBATCH -J MD5_checksum_verified_transfer | ||
| + | #SBATCH --mail-type=ALL | ||
| + | |||
| + | thefile=<GPFSpath> | ||
| + | storedfile=<HPSSpath> | ||
| + | |||
| + | # Generate checksum on fly (-c on) | ||
| + | hsi -q put -c on $thefile : $storedfile | ||
| + | pid=$! | ||
| − | + | # Check the exit code of the HSI process | |
| − | + | status=$? | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | if [ ! $status == 0 ]; then |
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| − | + | # verify checksum | |
| − | + | hsi lshash $storedfile | |
| − | + | status=$? | |
| − | + | ||
| − | + | if [ ! $status == 0 ]; then | |
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| − | + | # get the file back with checksum | |
| − | + | hsi get -c on $storedfile | |
| − | + | status=$? | |
| − | </ | + | |
| + | if [ ! $status == 0 ]; then | ||
| + | echo 'HSI returned non-zero code.' | ||
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
| + | fi | ||
| + | </source> | ||
| − | + | ==== Prior to HSI version 4.0.1.1 ==== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | This will checksum the contents of the HPSSpath against the original GPFSpath after the transfer has finished. | |
| − | < | + | <source lang="bash"> |
| − | #!/bin/ | + | #!/bin/bash |
| − | # | + | #SBATCH -t 72:00:00 |
| − | # | + | #SBATCH -p archivelong |
| − | # | + | #SBATCH -N 1 |
| − | # | + | #SBATCH -J checksum_verified_transfer |
| + | #SBATCH --mail-type=ALL | ||
| − | thefile=< | + | thefile=<GPFSpath> |
| − | storedfile=< | + | storedfile=<HPSSpath> |
# Generate checksum on fly using a named pipe so that file is only read from GPFS once | # Generate checksum on fly using a named pipe so that file is only read from GPFS once | ||
| Line 313: | Line 1,002: | ||
pid=$! | pid=$! | ||
md5sum /tmp/NPIPE |tee /tmp/$fname.md5 | md5sum /tmp/NPIPE |tee /tmp/$fname.md5 | ||
| − | rm | + | rm -f /tmp/NPIPE |
# Check the exit code of the HSI process | # Check the exit code of the HSI process | ||
wait $pid | wait $pid | ||
| − | + | status=$? | |
| − | if [ $ | + | |
| − | + | if [ ! $status == 0 ]; then | |
| − | + | echo 'HSI returned non-zero code.' | |
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
fi | fi | ||
| − | |||
# change filename to stdin in checksum file | # change filename to stdin in checksum file | ||
| Line 329: | Line 1,021: | ||
# verify checksum | # verify checksum | ||
hsi -q get - : $storedfile | md5sum -c /tmp/$fname.md5 | hsi -q get - : $storedfile | md5sum -c /tmp/$fname.md5 | ||
| − | + | status=$? | |
| − | if [ $ | + | |
| − | + | if [ ! $status == 0 ]; then | |
| − | + | echo 'HSI returned non-zero code.' | |
| + | /scinet/niagara/bin/exit2msg $status | ||
| + | exit $status | ||
| + | else | ||
| + | echo 'TRANSFER SUCCESSFUL' | ||
fi | fi | ||
| − | </ | + | </source> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | == ''' | + | == '''Access to HPSS using Globus''' == |
| − | + | * <font color=red> Please note that Globus access to HPSS is disabled until further notice, due to lack of version compatibility.</font> | |
| − | + | * You may now transfer data between SciNet's HPSS and an external source | |
| + | * Follow the link below | ||
| + | https://globus.computecanada.ca | ||
| + | : Enter your Compute Canada username and password. | ||
| + | * In the 'File Transfer' tab, enter ''''Compute Canada HPSS'''' as one of the Endpoints. To authenticate this endpoint, enter your SciNet username and password. | ||
| + | * You may read more about Compute Canada's Globus Portal here: | ||
| + | https://docs.computecanada.ca/wiki/Globus | ||
| − | + | == '''Access to HPSS using SME''' == | |
| − | * | + | * Storage Made Easy - SME - is an Enterprise Cloud Portal adopted by SciNet to allow our users to access HPSS |
| − | * | + | * Best suitable for light transfers to/from your personal computer and to navigate your contents on HPSS |
| − | + | * Follow the link below using a web browser and login with your SicNet UserID and password. Under File Manager you will find the "'''SciNet HPSS'''" folder. | |
| + | https://sme.scinet.utoronto.ca | ||
| + | * SME can be configured as a DropBox. To download the Free Cloud File Manager native to your OS (Windows, Mac, Linux, mobile), follow the link below: | ||
| + | https://www.storagemadeeasy.com/clients_and_tools/ | ||
| + | Once you have downloaded and installed the Cloud Manager App, fill up the following information: | ||
| + | Server location | ||
| + | https://sme.scinet.utoronto.ca/api | ||
| + | * You may learn more about SME capabilities and features here: | ||
| + | https://www.storagemadeeasy.com/ownFileserver/ | ||
| + | https://www.storagemadeeasy.com/pricing/#features (Enterprise) | ||
| + | https://storagemadeeasy.com/faq/ | ||
| − | == | + | == '''User provided Content/Suggestions''' == |
| − | + | == '''[[HPSS-by-pomes|Packing up large data sets and putting them on HPSS]]''' == | |
| − | + | (Pomés group recommendations) | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | [[Data Management|BACK TO Data Management]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 13:28, 9 August 2018
|
WARNING: SciNet is in the process of replacing this wiki with a new documentation site. For current information, please go to https://docs.scinet.utoronto.ca |
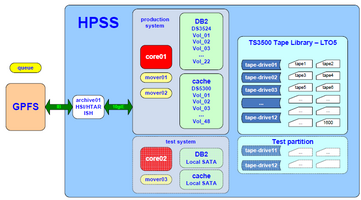
| Topology Overview | Submission Queue |
| Servers Rack | TS3500 Library |
High Performance Storage System
The High Performance Storage System (HPSS wikipedia) is a tape-backed hierarchical storage system that provides a significant portion of the allocated storage space at SciNet. It is a repository for archiving data that is not being actively used. Data can be returned to the active GPFS filesystem when it is needed.
Since this system is intended for large data storage, it is accessible only to groups who have been awarded storage space at SciNet beyond 5TB in the yearly RAC resource allocation round. However, upon request, any user may be awarded access to HPSS, up to 2TB per group, so that you may get familiar with the system (just email support@scinet.utoronto.ca)
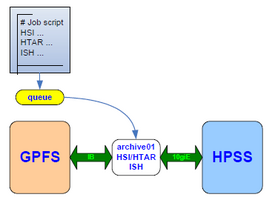
Access and transfer of data into and out of HPSS is done under the control of the user, whose interaction is expected to be scripted and submitted as a batch job, using one or more of the following utilities:
- HSI is a client with an ftp-like functionality which can be used to archive and retrieve large files. It is also useful for browsing the contents of HPSS.
- HTAR is a utility that creates tar formatted archives directly into HPSS. It also creates a separate index file (.idx) that can be accessed and browsed quickly.
- ISH is a TUI utility that can perform an inventory of the files and directories in your tarballs.
We're currently running HPSS v 7.3.3 patch 6, and HSI/HTAR version 4.0.1.2
Why should I use and trust HPSS?
- HPSS is a 25 year-old collaboration between IBM and the DoE labs in the US, and is used by about 45 facilities in the “Top 500” HPC list (plus some black-sites).
- Over 2.5 ExaBytes of combined storage world-wide.
- The top 3 sites in the World report (fall 2017) having 360PB, 220PB and 125PB in production (ECMWF, UKMO and BNL)
- Environment Canada also adopted HPSS in 2017 to store Nav Canada data as well as to serve as their own archive. Currently has 2 X 100PB capacity installed.
- The SciNet HPSS system has been providing nearline capacity for important research data in Canada since early 2011, already at 10PB levels in 2018
- Very reliable, data redundancy and data insurance built-in (dual copies of everything are kept on tapes at SciNet)
- Data on cache and tapes can be geo-distributed for further resilience and HA.
- Highly scalable; current performance at SciNet - after a modest upgrade in 2017 - Ingest: ~150 TB/day, Recall: ~45 TB/day (aggregated).
- HSI/HTAR clients also very reliable and used on several HPSS sites. ISH was written at SciNet.
- HPSS fits well with the Storage Capacity Expansion Plan at SciNet (pdf presentation)
Guidelines
- Expanded storage capacity is provided on tape -- a media that is not suited for storing small files. Files smaller than ~200MB should be grouped into tarballs with tar or htar.
- Optimal performance for aggregated transfers and allocation on tapes is obtained with tarballs of size 500GB or less, whether ingested by htar or hsi ( WHY?)
- We strongly urge that you use the sample scripts we are providing as the basis for your job submissions.
- Make sure to check the application's exit code and returned logs for errors after any data transfer or tarball creation process
New to the System?
The first step is to email scinet support and request an HPSS account (or else you will get "Error - authentication/initialization failed" and 71 exit codes).
THIS set of instructions on the wiki is the best and most compressed "manual" we have. It may seem a bit overwhelming at first, because of all the job script templates we make available below (they are here so you don't have to think too much, just copy and paste), but if you approach the index at the top as a "case switch" mechanism for what you intend to do, everything falls in place.
Try this sequence:
1) take a look around HPSS using an interactive HSI session
(most linux shell commands have an equivalent in HPSS)
2) archive a small test directory using HTAR
2a) use step 1) to see what happened
3a) use step 1) to see what happened
4) archive a small test directory using HSI
4a) use step 1) to see what happened
5) now try the other cases and so on. In a couple of hours you'll be in pretty good shape.
Bridge between BGQ and HPSS
At this time BGQ users will have to migrate data to Niagara scratch prior to transferring it to HPSS. We are looking for ways to improve this workflow.
Access Through the Queue System
All access to the archive system is done through the NIA queue system.
- Job submissions should be done to the 'archivelong' queue or the 'archiveshort'
- Short jobs are limited to 1H walltime by default. Long jobs (> 1H) are limited to 72H walltime.
- Users are limited to only 2 long jobs and 2 short jobs at the same time, and 10 jobs total on the each queue.
- There can only be 5 long jobs running at any given time overall. Remaining submissions will be placed on hold for the time being. So far we have not seen a need for overall limit on short jobs.
The status of pending jobs can be monitored with squeue specifying the archive queue:
squeue -p archiveshort OR squeue -p archivelong
Access Through an Interactive HSI session
- You may want to acquire an interactive shell, start an HSI session and navigate the archive naming-space. Keep in mind, you're restricted to 1H.
pinto@nia-login07:~$ salloc -p archiveshort -t 1:00:00 salloc: Granted job allocation 50918 salloc: Waiting for resource configuration salloc: Nodes hpss-archive02-ib are ready for job hpss-archive02-ib:~$ hpss-archive02-ib:~$ hsi (DON'T FORGET TO START HSI) ****************************************************************** * Welcome to HPSS@SciNet - High Perfomance Storage System * * * * INFO: THIS IS THE NEW 7.5.1 HPSS SYSTEM! * * * * Contact Information: support@scinet.utoronto.ca * * NOTE: do not transfer SMALL FILES with HSI. Use HTAR instead * * CHECK THE INTEGRITY OF YOUR TARBALLS * ****************************************************************** [HSI]/archive/s/scinet/pinto-> ls [HSI]/archive/s/scinet/pinto-> cd <some directory>
Scripted File Transfers
File transfers in and out of the HPSS should be scripted into jobs and submitted to the archivelong queue or the archiveshort . See generic example below:
<source lang="bash">
- !/bin/bash -l
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J htar_create_tarball_in_hpss
- SBATCH --mail-type=ALL
echo "Creating a htar of finished-job1/ directory tree into HPSS"
trap "echo 'Job script not completed';exit 129" TERM INT
- Note that your initial directory in HPSS will be $ARCHIVE
DEST=$ARCHIVE/finished-job1.tar
- htar WILL overwrite an existing file with the same name so check beforehand.
hsi ls $DEST &> /dev/null status=$?
if [ $status == 0 ]; then
echo 'File $DEST already exists. Nothing has been done' exit 1
fi
cd $SCRATCH/workarea/ htar -Humask=0137 -cpf $ARCHIVE/finished-job1.tar finished-job1/ status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HTAR returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source> Note: Always trap the execution of your jobs for abnormal terminations, and be sure to return the exit code
Job Dependencies
Typically data will be recalled to /scratch when it is needed for analysis. Job dependencies can be constructed so that analysis jobs wait in the queue for data recalls before starting. The qsub flag is
--dependency=<type:JOBID>
where JOBID is the job number of the archive recalling job that must finish successfully before the analysis job can start.
Here is a short cut for generating the dependency (lookup data-recall.sh samples):
hpss-archive02-ib:~$ sbatch $(sbatch data-recall.sh | awk {print "--dependency=afterany:"$1}') job-to-work-on-recalled-data.sh
HTAR
Please aggregate small files (<~200MB) into tarballs or htar files.
Keep your tarballs to size 500GB or less, whether ingested by htar or hsi ( WHY?)
HTAR is a utility that is used for aggregating a set of files and directories, by using a sophisticated multithreaded buffering scheme to write files directly from GPFS into HPSS, creating an archive file that conforms to the POSIX TAR specification, thereby achieving a high rate of performance. HTAR does not do gzip compression, however it already has a built-in checksum algorithm.
Caution
- Files larger than 68 GB cannot be stored in an HTAR archive. If you attempt to start a transfer with any files larger than 68GB the whole HTAR session will fail, and you'll get a notification listing all those files, so that you can transfer them with HSI.
- Files with pathnames too long will be skipped (greater than 100 characters), so as to conform with TAR protocol (POSIX 1003.1 USTAR) -- Note that the HTAR will erroneously indicate success, however will produce exit code 70. For now, you can check for this type of error by "grep Warning my.output" after the job has completed.
- Unlike with cput/cget in HSI, "prompt before overwrite", this is not the default with (h)tar. Be careful not to unintentionally overwrite a previous htar destination file in HPSS. There could be a similar situation when extracting material back into GPFS and overwriting the originals. Be sure to double-check the logic in your scripts.
- Check the HTAR exit code and log file before removing any files from the GPFS active filesystems.
=== HTAR Usage ===
- To write the file1 and file2 files to a new archive called files.tar in the default HPSS home directory, and preserve mask attributes (-p), enter:
htar -cpf files.tar file1 file2
OR
htar -cpf $ARCHIVE/files.tar file1 file2
- To write a subdirA to a new archive called subdirA.tar in the default HPSS home directory, enter:
htar -cpf subdirA.tar subdirA
- To extract all files from the archive file called proj1.tar in HPSS into the project1/src directory in GPFS, and use the time of extraction as the modification time, enter:
cd project1/src
htar -xpmf proj1.tar
- To display the names of the files in the out.tar archive file within the HPSS home directory, enter (the out.tar.idx file will be queried):
htar -vtf out.tar
- To ensure that both the htar and the .idx files have read permissions to other members in your group use the umask option
htar -Humask=0137 ....
For more details please check the HTAR - Introduction or the HTAR Man Page online
Sample tarball create
<source lang="bash">
- !/bin/bash -l
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J htar_create_tarball_in_hpss
- SBATCH --mail-type=ALL
trap "echo 'Job script not completed';exit 129" TERM INT
- Note that your initial directory in HPSS will be /archive/$(id -gn)/$(whoami)/
DEST=$ARCHIVE/finished-job1.tar
- htar WILL overwrite an existing file with the same name so check beforehand.
hsi ls $DEST &> /dev/null status=$?
if [ $status == 0 ]; then
echo 'File $DEST already exists. Nothing has been done' exit 1
fi
cd $SCRATCH/workarea/ htar -Humask=0137 -cpf $DEST finished-job1/ status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HTAR returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Note: If you attempt to start a transfer with any files larger than 68GB the whole HTAR session will fail, and you'll get a notification listing all those files, so that you can transfer them with HSI.
---------------------------------------- INFO: File too large for htar to handle: finished-job1/file1 (86567185745 bytes) INFO: File too large for htar to handle: finished-job1/file2 (71857244579 bytes) ERROR: 2 oversize member files found - please correct and retry ERROR: [FATAL] error(s) generating filename list HTAR: HTAR FAILED ###WARNING htar returned non-zero exit status
Sample tarball list
<source lang="bash">
- !/bin/bash -l
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J htar_list_tarball_in_hpss
- SBATCH --mail-type=ALL
trap "echo 'Job script not completed';exit 129" TERM INT
- Note that your initial directory in HPSS will be $ARCHIVE
DEST=$ARCHIVE/finished-job1.tar
htar -tvf $DEST status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HTAR returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Sample tarball extract
<source lang="bash">
- !/bin/bash
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J htar_extract_tarball_from_hpss
- SBATCH --mail-type=ALL
trap "echo 'Job script not completed';exit 129" TERM INT
- Note that your initial directory in HPSS will be $ARCHIVE
cd $SCRATCH/recalled-from-hpss htar -xpmf $ARCHIVE/finished-job1.tar status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HTAR returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
HSI
HSI may be the primary client with which some users will interact with HPSS. It provides an ftp-like interface for archiving and retrieving tarballs or directory trees. In addition it provides a number of shell-like commands that are useful for examining and manipulating the contents in HPSS. The most commonly used commands will be:
| cput | Conditionally saves or replaces a HPSSpath file to GPFSpath if the GPFS version is new or has been updated
cput [options] GPFSpath [: HPSSpath] |
| cget | Conditionally retrieves a copy of a file from HPSS to GPFS only if a GPFS version does not already exist.
cget [options] [GPFSpath :] HPSSpath |
| cd,mkdir,ls,rm,mv | Operate as one would expect on the contents of HPSS. |
| lcd,lls | Local commands to GPFS |
- There are 3 distinctions about HSI that you should keep in mind, and that can generate a bit of confusion when you're first learning how to use it:
- HSI doesn't currently support renaming directories paths during transfers on-the-fly, therefore the syntax for cput/cget may not work as one would expect in some scenarios, requiring some workarounds.
- HSI has an operator ":" which separates the GPFSpath and HPSSpath, and must be surrounded by whitespace (one or more space characters)
- The order for referring to files in HSI syntax is different from FTP. In HSI the general format is always the same, GPFS first, HPSS second, cput or cget:
GPFSfile : HPSSfile
For example, when using HSI to store the tarball file from GPFS into HPSS, then recall it to GPFS, the following commands could be used:
cput tarball-in-GPFS : tarball-in-HPSS
cget tarball-recalled : tarball-in-HPSS
unlike with FTP, where the following syntax would be used:
put tarball-in-GPFS tarball-in-HPSS
get tarball-in-HPSS tarball-recalled
- Simple commands can be executed on a single line.
hsi "mkdir LargeFilesDir; cd LargeFilesDir; cput tarball-in-GPFS : tarball-in-HPSS"
- More complex sequences can be performed using an except such as this:
hsi <<EOF
mkdir LargeFilesDir
cd LargeFilesDir
cput tarball-in-GPFS : tarball-in-HPSS
lcd $SCRATCH/LargeFilesDir2/
cput -Ruph *
end
EOF
- The commands below are equivalent, but we recommend that you always use full path, and organize the contents of HPSS, where the default HSI directory placement is $ARCHIVE:
hsi cput tarball
hsi cput tarball : tarball
hsi cput $SCRATCH/tarball : $ARCHIVE/tarball
- There are no known issues renaming files on-the-fly:
hsi cput $SCRATCH/tarball1 : $ARCHIVE/tarball2
hsi cget $SCRATCH/tarball3 : $ARCHIVE/tarball2
- However the syntax forms such as the ones below will fail, since they rename the directory paths.
hsi cput -Ruph $SCRATCH/LargeFilesDir : $ARCHIVE/LargeFilesDir (FAILS) OR hsi cget -Ruph $SCRATCH/LargeFilesDir : $ARCHIVE/LargeFilesDir2 (FAILS) OR hsi cput -Ruph $SCRATCH/LargeFilesDir/* : $ARCHIVE/LargeFilesDir2 (FAILS) OR hsi cget -Ruph $SCRATCH/LargeFilesDir : $ARCHIVE/LargeFilesDir (FAILS)
One workaround is the following 2-steps process, where you do a "lcd " in GPFS first, and recursively transfer the whole directory (-R), keeping the same name. You may use '-u' option to resume a previously disrupted session, and the '-p' to preserve timestamp, and '-h' to keep the links.
hsi <<EOF
lcd $SCRATCH
cget -Ruph LargeFilesDir
end
EOF
Another workaround is do a "lcd" into the GPFSpath first and a "cd" in the HPSSpath, but transfer the files individually with the '*' wild character. This option lets you change the directory name:
hsi <<EOF
lcd $SCRATCH/LargeFilesDir
mkdir $ARCHIVE/LargeFilesDir2
cd $ARCHIVE/LargeFilesDir2
cput -Ruph *
end
EOF
Documentation
Complete documentation on HSI is available from the Gleicher Enterprises links below. You may peruse those links and come with alternative syntax forms. You may even be already familiar with HPSS/HSI from other HPC facilities, that may or not have procedures similar to ours. HSI doesn't always work as expected when you go outside of our recommended syntax, so we strongly urge that you use the sample scripts we are providing as the basis for your job submissions
Note: HSI returns the highest-numbered exit code, in case of multiple operations in the same hsi session. You may use '/scinet/niagara/bin/exit2msg $status' to translate those codes into intelligible messages
Typical Usage Scripts
The most common interactions will be putting data into HPSS, examining the contents (ls,ish), and getting data back onto GPFS for inspection or analysis.
Sample data offload
<source lang="bash">
- !/bin/bash
- This script is named: data-offload.sh
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J offload
- SBATCH --mail-type=ALL
trap "echo 'Job script not completed';exit 129" TERM INT
- individual tarballs already exist
/usr/local/bin/hsi -v <<EOF1 mkdir put-away cd put-away cput $SCRATCH/workarea/finished-job1.tar.gz : finished-job1.tar.gz end EOF1 status=$? if [ ! $status == 0 ];then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi
/usr/local/bin/hsi -v <<EOF2 mkdir put-away cd put-away cput $SCRATCH/workarea/finished-job2.tar.gz : finished-job2.tar.gz end EOF2 status=$? if [ ! $status == 0 ];then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi
trap - TERM INT </source>
Note: as in the above example, we recommend that you capture the (highest-numbered) exit code for each hsi session independently. And remember, you may improve your exit code verbosity by adding the excerpt below to your scripts:
<source lang="bash">
if [ ! $status == 0 ];then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Sample data list
A very trivial way to list the contents of HPSS would be to just submit the HSI 'ls' command. <source lang="bash">
- !/bin/bash
- This script is named: data-list.sh
- SBATCH -t 1:00:00
- SBATCH -p archiveshort
- SBATCH -N 1
- SBATCH -J hpss_ls
- SBATCH --mail-type=ALL
/usr/local/bin/hsi -v <<EOF cd put-away ls -R end EOF </source> Warning: if you have a lot of files, the ls command will take a long time to complete. For instance, about 400,000 files can be listed in about an hour. Adjust the walltime accordingly, and be on the safe side.
However, we provide a much more useful and convenient way to explore the contents of HPSS with the inventory shell ISH. This example creates an index of all the files in a user's portion of the namespace. The list is placed in the directory /home/$(whoami)/.ish_register that can be inspected from the login nodes. <source lang="bash">
- !/bin/bash
- This script is named: data-list.sh
- SBATCH -t 1:00:00
- SBATCH -p archiveshort
- SBATCH -N 1
- SBATCH -J hpss_index
- SBATCH --mail-type=ALL
INDEX_DIR=$HOME/.ish_register if ! [ -e "$INDEX_DIR" ]; then
mkdir -p $INDEX_DIR
fi
export ISHREGISTER="$INDEX_DIR" /scinet/niagara/bin/ish hindex </source> Note: the above warning on collecting the listing for many files applies here too.
This index can be browsed or searched with ISH on the development nodes. <source lang="bash"> hpss-archive02-ib:~$ /scinet/niagara/bin/ish ~/.ish_register/hpss.igz [ish]hpss.igz> help </source>
ISH is a powerful tool that is also useful for creating and browsing indices of tar and htar archives, so please look at the documentation or built in help.
Sample data recall
<source lang="bash">
- !/bin/bash
- This script is named: data-recall.sh
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J recall_files
- SBATCH --mail-type=ALL
trap "echo 'Job script not completed';exit 129" TERM INT
mkdir -p $SCRATCH/recalled-from-hpss
- individual tarballs previously organized in HPSS inside the put-away-on-2010/ folder
hsi -v << EOF cget $SCRATCH/recalled-from-hpss/Jan-2010-jobs.tar.gz : $ARCHIVE/put-away-on-2010/Jan-2010-jobs.tar.gz cget $SCRATCH/recalled-from-hpss/Feb-2010-jobs.tar.gz : $ARCHIVE/put-away-on-2010/Feb-2010-jobs.tar.gz end EOF status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagar/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
We should emphasize that a single cget of multiple files (rather than several separate gets) allows HSI to do optimization, as in the following example:
<source lang="bash">
- !/bin/bash
- This script is named: data-recall.sh
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J recall_files_optimized
- SBATCH --mail-type=AL
trap "echo 'Job script not completed';exit 129" TERM INT mkdir -p $SCRATCH/recalled-from-hpss
- individual tarballs previously organized in HPSS inside the put-away-on-2010/ folder
hsi -v << EOF lcd $SCRATCH/recalled-from-hpss/ cd $ARCHIVE/put-away-on-2010/ cget Jan-2010-jobs.tar.gz Feb-2010-jobs.tar.gz end EOF status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Sample transferring directories
Remember, it's not possible to rename directories on-the-fly:
hsi cget -Ruph $SCRATCH/LargeFiles-recalled : $ARCHIVE/LargeFiles (FAILS)
One workaround is transfer the whole directory (and sub-directories) recursively: <source lang="bash">
- !/bin/bash
- This script is named: data-recall.sh
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J recall_directories
- SBATCH --mail-type=ALL
trap "echo 'Job script not completed';exit 129" TERM INT
mkdir -p $SCRATCH/recalled
hsi -v << EOF lcd $SCRATCH/recalled cd $ARCHIVE/ cget -Ruph LargeFiles end EOF status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Another workaround is to transfer files and subdirectories individually with the "*" wild character:
<source lang="bash">
- !/bin/bash
- This script is named: data-recall.sh
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J recall_directories
- SBATCH --mail-type=AL
trap "echo 'Job script not completed';exit 129" TERM INT
mkdir -p $SCRATCH/LargeFiles-recalled
hsi -v << EOF lcd $SCRATCH/LargeFiles-recalled cd $ARCHIVE/LargeFiles cget -Ruph * end EOF status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
- For more details please check the HSI Introduction, the HSI Man Page or the or the hsi help
ISH
Documentation and Usage
File and directory management
Moving/renaming
- you may use 'mv' or 'cp' in the same way as the linux version.
<source lang="bash">
- !/bin/bash
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J deletion_script
- SBATCH --mail-type=ALL
echo "HPSS file and directory management"
trap "echo 'Job script not completed';exit 129" TERM INT
/usr/local/bin/hsi -v <<EOF1
mkdir $ARCHIVE/2011 mv $ARCHIVE/oldjobs $ARCHIVE/2011 cp -r $ARCHIVE/almostfinished/*done $ARCHIVE/2011
end EOF1 status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Deletions
Recommendations
- Be careful with the use of 'cd' commands to non-existing directories before the 'rm' command. Results may be unpredictable
- Avoid the use of the stand alone wild character *. If necessary, whenever possible have it bound to common patterns, such as '*.tmp', so to limit unintentional mis-happens
- Avoid using relative paths, even the env variable $ARCHIVE. Better to explicitly expand the full paths in your scripts
- Avoid using recursive/looped deletion instructions on $SCRATCH contents from the archive job scripts. Even on $ARCHIVE contents, it may be better to do it as an independent job submission, after you have verified that the original ingestion into HPSS finished without any issues.
Typical example
<source lang="bash">
- !/bin/bash
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J deletion_script
- SBATCH --mail-type=ALL
echo "Deletion of an outdated directory tree into HPSS"
trap "echo 'Job script not completed';exit 129" TERM INT
- Note that the initial directory in HPSS ($ARCHIVE) has the path explicitly expanded
/usr/local/bin/hsi -v <<EOF1
rm /archive/s/scinet/pinto/*.tmp rm -R /archive/s/scinet/pinto/obsolete
end EOF1 status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Deleting with an interactive HSI session
- You may feel more comfortable acquiring an interactive shell, starting an HSI session and proceeding with your deletions that way. Keep in mind, you're restricted to 1H.
- After using the qsub -q archive -I command you'll get a standard shell prompt on an archive execution node (hpss-archive02), as you would on any compute node. However you will need to run HSI or HTAR to access resources on HPSS.
- HSI will give you a prompt very similar to a standard shell, where your can navigate around using commands such 'ls', 'cd', 'pwd', etc ... NOTE: not every bash command has an equivalent on HSI - for instance, you can not 'vi' or 'cat'.
pinto@nia-login07:~$ salloc -p archiveshort -t 1:00:00 salloc: Granted job allocation 50359 salloc: Waiting for resource configuration salloc: Nodes hpss-archive02-ib are ready for job hpss-archive02-ib:~$ hsi ****************************************************************** * Welcome to HPSS@SciNet - High Perfomance Storage System * * * * INFO: THIS IS THE NEW 7.5.1 HPSS SYSTEM! * * * * Contact Information: support@scinet.utoronto.ca * * NOTE: do not transfer SMALL FILES with HSI. Use HTAR instead * * CHECK THE INTEGRITY OF YOUR TARBALLS * ****************************************************************** [HSI]/archive/s/scinet/pinto-> rm -R junk
HPSS for the 'Watchmaker'
Efficient alternative to htar
<source lang="bash">
- !/bin/bash
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J tar_create_tarball_in_hpss_with_hsi_by_piping
- SBATCH --mail-type=ALL
trap "echo 'Job script not completed';exit 129" TERM INT
- Note that your initial directory in HPSS will be $ARCHIVE
- When using a pipeline like this
set -o pipefail
- to put (cput will fail)
tar -c $SCRATCH/mydir | hsi put - : $ARCHIVE/mydir.tar status=$? if [ ! $status == 0 ]; then
echo 'TAR+HSI+piping returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi
- to immediately generate an index
ish hindex $ARCHIVE/mydir.tar status=$? if [ ! $status == 0 ]; then
echo 'ISH returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi
- to get
- cd $SCRATCH
- hsi cget - : $ARCHIVE/mydir.tar | tar -xv
- status=$?
- if [ ! $status == 0 ]; then
- echo 'TAR+HSI+piping returned non-zero code.'
- /scinet/niagara/bin/exit2msg $status
- exit $status
- else
- echo 'TRANSFER SUCCESSFUL'
- fi
trap - TERM INT
</source> Notes:
- Combining commands in this fashion, besides being HPSS-friendly, should not be that noticeably slower than the recursive put with HSI that stores each file one by one. However, reading the files back from tape in this format will be many times faster. It would also overcome the current 68GB limit on the size of stored files that we have with htar.
- To top things off, we recommend indexing with ish (in the same script) immediately after the tarball creation , while it resides in the HPSS cache. It would be as if htar was used.
- To ensure that an error at any stage of the pipeline shows up in the returned status use: set -o pipefail (The default is to return the status of the last command in the pipeline and this is not what you want.)
- Optimal performance for aggregated transfers and allocation on tapes is obtained with tarballs of size 500GB or less, whether ingested by htar or hsi ( WHY?). Be sure to check the contents of the directory tree with 'du' for the total amount of data before sending them to the tar+HSI piping.
Multi-threaded gzip'ed compression with pigz
We compiled multi-threaded implementation of gzip called pigz (http://zlib.net/pigz/). It's now part of the "extras" module. It can also be used on any compute or devel nodes. This makes the execution of the previous version of the script much quicker than if you were to use 'tar -cfz'. In addition, by piggy-backing ISH to the end of the script, it will know what to do with the just created mydir.tar.gz compressed tarball.
<source lang="bash">
- !/bin/bash
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J tar_create_compressed_tarball_in_hpss_with_hsi_by_piping
- SBATCH --mail-type=ALL
trap "echo 'Job script not completed';exit 129" TERM INT
- Note that your initial directory in HPSS will be $ARCHIVE
- When using a pipeline like this
set -o pipefail
load module extras
- to put (cput will fail)
tar -c $SCRATCH/mydir | pigz | hsi put - : $ARCHIVE/mydir.tar.gz status=$? if [ ! $status == 0 ]; then
echo 'TAR+PIGZ+HSI+piping returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Content Verification
HTAR CRC checksums
Specifies that HTAR should generate CRC checksums when creating the archive.
<source lang="bash">
- !/bin/bash
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J htar_create_tarball_in_hpss_with_checksum_verification
- SBATCH --mail-type=ALL
trap "echo 'Job script not completed';exit 129" TERM INT
- Note that your initial directory in HPSS will be $ARCHIVE
cd $SCRATCH/workarea
- to put
htar -Humask=0137 -cpf $ARCHIVE/finished-job1.tar -Hcrc -Hverify=1 finished-job1/
- to get
- mkdir $SCRATCH/verification
- cd $SCRATCH/verification
- htar -Hcrc -xvpmf $ARCHIVE/finished-job1.tar
status=$?
trap - TERM INT
if [ ! $status == 0 ]; then
echo 'HTAR returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Current HSI version - Checksum built-in
MD5 is the standard Hashing Algorithm for the HSI build at SciNet. For hsi ingestions with the '-c on' option you should be able to query the md5 hash with the hsi command 'hashli'. That value is stored as an UDA (User Defined Attribute) for each file (a feature of HPSS starting with 7.4)
The checksum algorithm is very CPU-intensive. Although the checksum code is compiled with a high level of compiler optimization, transfer rates can be significantly reduced when checksum creation or verification is in effect. The amount of degradation in transfer rates depends on several factors, such as processor speed, network transfer speed, and speed of the local filesystem (GPFS).
<source lang="bash">
- !/bin/bash
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J MD5_checksum_verified_transfer
- SBATCH --mail-type=ALL
thefile=<GPFSpath> storedfile=<HPSSpath>
- Generate checksum on fly (-c on)
hsi -q put -c on $thefile : $storedfile pid=$!
- Check the exit code of the HSI process
status=$?
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi
- verify checksum
hsi lshash $storedfile status=$?
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi
- get the file back with checksum
hsi get -c on $storedfile status=$?
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Prior to HSI version 4.0.1.1
This will checksum the contents of the HPSSpath against the original GPFSpath after the transfer has finished. <source lang="bash">
- !/bin/bash
- SBATCH -t 72:00:00
- SBATCH -p archivelong
- SBATCH -N 1
- SBATCH -J checksum_verified_transfer
- SBATCH --mail-type=ALL
thefile=<GPFSpath> storedfile=<HPSSpath>
- Generate checksum on fly using a named pipe so that file is only read from GPFS once
mkfifo /tmp/NPIPE cat $thefile | tee /tmp/NPIPE | hsi -q put - : $storedfile & pid=$! md5sum /tmp/NPIPE |tee /tmp/$fname.md5 rm -f /tmp/NPIPE
- Check the exit code of the HSI process
wait $pid status=$?
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi
- change filename to stdin in checksum file
sed -i.1 "s+/tmp/NPIPE+-+" /tmp/$fname.md5
- verify checksum
hsi -q get - : $storedfile | md5sum -c /tmp/$fname.md5 status=$?
if [ ! $status == 0 ]; then
echo 'HSI returned non-zero code.' /scinet/niagara/bin/exit2msg $status exit $status
else
echo 'TRANSFER SUCCESSFUL'
fi </source>
Access to HPSS using Globus
- Please note that Globus access to HPSS is disabled until further notice, due to lack of version compatibility.
- You may now transfer data between SciNet's HPSS and an external source
- Follow the link below
https://globus.computecanada.ca
- Enter your Compute Canada username and password.
- In the 'File Transfer' tab, enter 'Compute Canada HPSS' as one of the Endpoints. To authenticate this endpoint, enter your SciNet username and password.
- You may read more about Compute Canada's Globus Portal here:
https://docs.computecanada.ca/wiki/Globus
Access to HPSS using SME
- Storage Made Easy - SME - is an Enterprise Cloud Portal adopted by SciNet to allow our users to access HPSS
- Best suitable for light transfers to/from your personal computer and to navigate your contents on HPSS
- Follow the link below using a web browser and login with your SicNet UserID and password. Under File Manager you will find the "SciNet HPSS" folder.
https://sme.scinet.utoronto.ca
- SME can be configured as a DropBox. To download the Free Cloud File Manager native to your OS (Windows, Mac, Linux, mobile), follow the link below:
https://www.storagemadeeasy.com/clients_and_tools/
Once you have downloaded and installed the Cloud Manager App, fill up the following information:
Server location https://sme.scinet.utoronto.ca/api
- You may learn more about SME capabilities and features here:
https://www.storagemadeeasy.com/ownFileserver/ https://www.storagemadeeasy.com/pricing/#features (Enterprise) https://storagemadeeasy.com/faq/
User provided Content/Suggestions
Packing up large data sets and putting them on HPSS
(Pomés group recommendations)