User Codes
Astrophysics
Athena (explicit, uniform grid MHD code)
Athena is a straightforward C code which doesn't use a lot of libraries so it is pretty straightforward to build and compile on new machines.
It encapsulates its compiler flags, etc in an Makeoptions.in file which is then processed by configure. I've used the following additions to Makeoptions.in on TCS and GPC:
<source lang="sh"> ifeq ($(MACHINE),scinettcs)
CC = mpcc_r LDR = mpcc_r OPT = -O5 -q64 -qarch=pwr6 -qtune=pwr6 -qcache=auto -qlargepage -qstrict MPIINC = MPILIB = CFLAGS = $(OPT) LIB = -ldl -lm
else ifeq ($(MACHINE),scinetgpc)
CC = mpicc LDR = mpicc OPT = -O3 MPIINC = MPILIB = CFLAGS = $(OPT) LIB = -lm
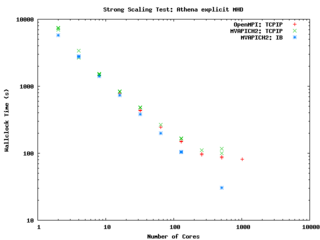
else ... endif endif </source> It performs quite well on the GPC, scaling extremely well even on a strong scaling test out to about 256 cores (32 nodes) on Gigabit ethernet, and performing beautifully on InfiniBand out to 512 cores (64 nodes).
-- ljdursi 19:20, 13 August 2009 (UTC)
FLASH3 (Adaptive Mesh reactive hydrodynamics; explict hydro/MHD)
FLASH encapsulates its machine-dependant information in the FLASH3/sites directory. For the GPC, you'll have to
module load intel module load openmpi module load hdf5/183-v16-openmpi
and with that, the following file (sites/scinetgpc/Makefile.h) works for me: <source lang="sh">
- Must do module load hdf5/183-v16-openmpi
HDF5_PATH = ${SCINET_HDF5_BASE} ZLIB_PATH = /usr/local
- ----------------------------------------------------------------------------
- Compiler and linker commands
- We use the f90 compiler as the linker, so some C libraries may explicitly
- need to be added into the link line.
- ----------------------------------------------------------------------------
- modules will put the right mpi in our path
FCOMP = mpif77 CCOMP = mpicc CPPCOMP = mpiCC LINK = mpif77
- ----------------------------------------------------------------------------
- Compilation flags
- Three sets of compilation/linking flags are defined: one for optimized
- code, one for testing, and one for debugging. The default is to use the
- _OPT version. Specifying -debug to setup will pick the _DEBUG version,
- these should enable bounds checking. Specifying -test is used for
- flash_test, and is set for quick code generation, and (sometimes)
- profiling. The Makefile generated by setup will assign the generic token
- (ex. FFLAGS) to the proper set of flags (ex. FFLAGS_OPT).
- ----------------------------------------------------------------------------
FFLAGS_OPT = -c -r8 -i4 -O3 -xSSE4.2 FFLAGS_DEBUG = -c -g -r8 -i4 -O0 FFLAGS_TEST = -c -r8 -i4
- if we are using HDF5, we need to specify the path to the include files
CFLAGS_HDF5 = -I${HDF5_PATH}/include
CFLAGS_OPT = -c -O3 -xSSE4.2 CFLAGS_TEST = -c -O2 CFLAGS_DEBUG = -c -g
MDEFS =
.SUFFIXES: .o .c .f .F .h .fh .F90 .f90
- ----------------------------------------------------------------------------
- Linker flags
- There is a seperate version of the linker flags for each of the _OPT,
- _DEBUG, and _TEST cases.
- ----------------------------------------------------------------------------
LFLAGS_OPT = -o LFLAGS_TEST = -o LFLAGS_DEBUG = -g -o
MACHOBJ =
MV = mv -f
AR = ar -r
RM = rm -f
CD = cd
RL = ranlib
ECHO = echo
</source>
-- ljdursi 22:11, 13 August 2009 (UTC)
Aeronautics
Chemistry
GAMESS (US)
The GAMESS version January 12, 2009 R3 was built using the Intel v11.1 compilers and v3.2.2 MPI library, according to the instructions in http://software.intel.com/en-us/articles/building-gamess-with-intel-compilers-intel-mkl-and-intel-mpi-on-linux/
The required build scripts - comp, compall, lked - and run script - rungms - were modified to account for our own installation. In order to build GAMESS one first must ensure that the intel and intelmpi modules are loaded ("module load intel intelmpi"). This applies to running GAMESS as well. The module "gamess" must also be loaded in order to run GAMESS ("module load gamess").
The modified scripts are in the file /scinet/gpc/src/gamess-on-scinet.tar.gz
Running GAMESS
- Make sure the directory /scratch/$USER/gamess-scratch exists (the $SCINET_RUNGMS script will create it if it does not exist)
- Make sure the modules: intel, intelmpi, gamess are loaded (in your .bashrc: "module load intel intelmpi gamess").
- Create a torque script to run GAMESS. Here is an example:
- The GAMESS executable is in $SCINET_GAMESS_HOME/gamess.00.x - The rungms script is in $SCINET_GAMESS_HOME/rungms (actually it is $SCINET_RUNGMS)
- For IB multinode runs, use the $SCINET_RUNGMS_IB script
- The rungms script takes 4 arguments: input file, executable number, number of compute processes, processors per node
For example, in order to run with the input file /scratch/$USER/gamesstest01, on 8 cpus, and the default version (00) of the executable on a machine with 8 cores:
# load the gamess module in .bashrc module load gamess
# run the program $SCINET_RUNGMS /scratch/$USER/gamesstest01 00 8 8
Here is a sample torque script for running a GAMESS calculation, on a single 8-core node:
<source lang="bash">
- !/bin/bash
- PBS -l nodes=1:ppn=8,walltime=48:00:00,os=centos53computeA
- PBS -N gamessjob
- To submit type: qsub gms.sh
- If not an interactive job (i.e. -I), then cd into the directory where
- I typed qsub.
if [ "$PBS_ENVIRONMENT" != "PBS_INTERACTIVE" ]; then
if [ -n "$PBS_O_WORKDIR" ]; then
cd $PBS_O_WORKDIR
fi
fi
- the input file is typically named something like "gamesjob.inp"
- so the script will be run like "$SCINET_RUNGMS gamessjob 00 8 8"
- load the gamess module if not in .bashrc already
- actually, it MUST be in .bashrc
- module load gamess
- run the program
$SCINET_RUNGMS gamessjob 00 8 8 </source>
Here is a similar script, but this one uses 2 InfiniBand-connected nodes, and runs the appropriate $SCINET_RUNGMS_IB script to actually run the job:
<source lang="bash">
- !/bin/bash
- PBS -l nodes=2:ib:ppn=8,walltime=48:00:00,os=centos53computeA
- PBS -N gamessjob
- To submit type: qsub gmsib.sh
- If not an interactive job (i.e. -I), then cd into the directory where
- I typed qsub.
if [ "$PBS_ENVIRONMENT" != "PBS_INTERACTIVE" ]; then
if [ -n "$PBS_O_WORKDIR" ]; then
cd $PBS_O_WORKDIR
fi
fi
- the input file is typically named something like "gamesjob.inp"
- so the script will be run like "$SCINET_RUNGMS gamessjob 00 8 8"
- load the gamess module if not in .bashrc already
- actually, it MUST be in .bashrc
- module load gamess
- This script requests InfiniBand-connected nodes (:ib above)
- so it must run with the IB version of the rungms script,
- $SCINET_RUNGMS_IB
$SCINET_RUNGMS_IB gamessjob 00 16 8 </source>
-- dgruner 5 October 2009
Tips from the Fekl Lab
Through trial and error, we have found a few useful things that we would like to share:
1. Two very useful, open-source programs for visualization of output files from GAMESS(US) and for generation of input files are MacMolPltand Avogadro. The are available for UNIX/LINUX, Windows and Mac based machines, HOWEVER: any input files that we have generated with these programs on a Windows-based machine do not run on Mac based machines. We don't know why.
2. WinSCP is a very useful tool that has a graphical user interface for moving files from a local machine to SCINET and vice versa. It also has text editing capabilities.
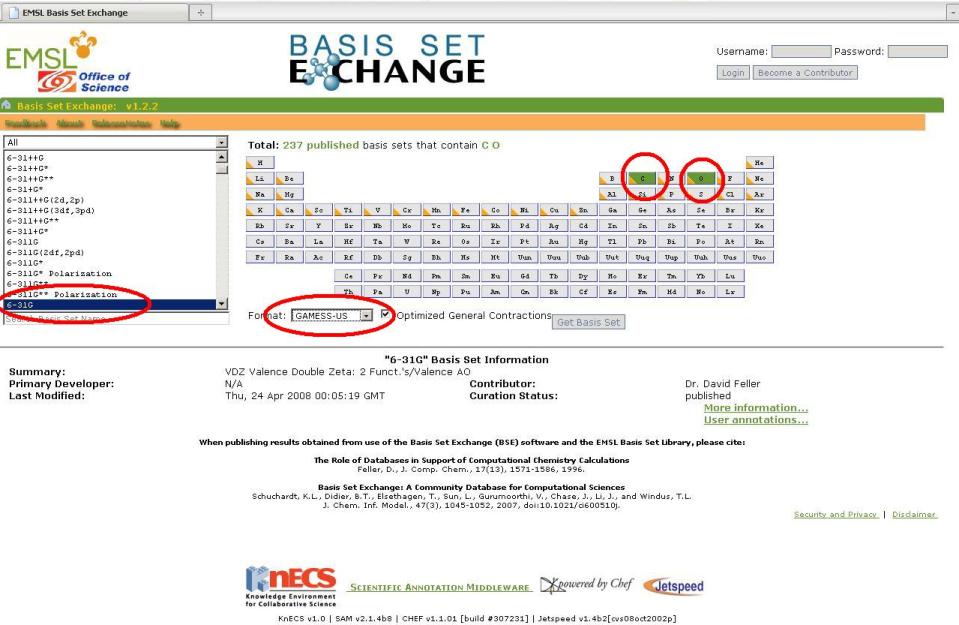
3. The ESML Basis Set Exchange is an excellent source for custom basis set or effective core potential parameters. Make sure that you specify "Gamess-US" in the format drop-down box.
4. The commercial program ChemCraft is a highly useful visualization program that has the ability to edit molecules in a very similar fashion to GaussView. It can also be customized to build GAMESS(US) input files.
Anatomy of a GAMESS(US) Input File
$CONTRL SCFTYP=RHF RUNTYP=OPTIMIZE DFTTYP=M06-L MAXIT=199 MULT=1 NOSYM=1
ECP=READ $END
$SYSTEM TIMLIM=525600 MWORDS=1750 PARALL=.TRUE. $END
$BASIS GBASIS=CUSTOMNI EXTFIL=.t. $END
$SCF DIRSCF=.TRUE. FDIFF=.f. $END
$STATPT OPTTOL=0.0001 NSTEP=500 HSSEND=.t. $END
$DATA
Mo_BDT3
C1
MOLYBDENUM 42.0 5.7556500000 4.4039600000 16.5808400000
SULFUR 16.0 7.4169700000 3.1956300000 15.2089300000
SULFUR 16.0 4.0966800000 3.2258300000 15.1761100000
SULFUR 16.0 3.9677300000 4.4940500000 18.3266100000
SULFUR 16.0 7.1776900000 3.5815000000 18.4485200000
SULFUR 16.0 4.3776600000 6.2447400000 15.6786900000
SULFUR 16.0 7.5478700000 6.0679800000 16.2223700000
CARBON 6.0 6.4716900000 2.1004800000 14.1902300000
CARBON 6.0 5.0690300000 2.1781400000 14.1080700000
CARBON 6.0 4.8421800000 4.2701300000 19.8855500000
CARBON 6.0 6.1969000000 3.9249600000 19.9397400000
CARBON 6.0 6.8280600000 3.7834200000 21.1913200000
CARBON 6.0 5.7697600000 7.6933500000 17.4241800000
CARBON 6.0 7.2043100000 7.9413600000 17.8281100000
CARBON 6.0 5.5051400000 7.0409700000 14.5903800000
CARBON 6.0 6.8905200000 6.9194700000 14.7626200000
CARBON 6.0 7.7396400000 7.5379800000 13.8285700000
HYDROGEN 1.0 8.8190700000 7.4520600000 13.9252200000
CARBON 6.0 7.2169400000 8.2960300000 12.7704100000
HYDROGEN 1.0 7.8667000000 8.7825100000 12.0575600000
CARBON 6.0 5.8260300000 8.4502300000 12.6467800000
HYDROGEN 1.0 5.4143000000 9.0544300000 11.8493100000
CARBON 6.0 4.9881500000 7.8192300000 13.5528400000
HYDROGEN 1.0 3.9090500000 7.9420000000 13.4583700000
CARBON 6.0 7.1538500000 1.1569600000 13.4143900000
CARBON 6.0 4.4018100000 1.3603900000 13.1919900000
CARBON 6.0 6.4791600000 0.3185500000 12.5353300000
CARBON 6.0 5.0837400000 0.4369500000 12.4084900000
HYDROGEN 1.0 7.0116000000 -0.4099400000 11.9434600000
HYDROGEN 1.0 8.2399000000 1.0702400000 13.4937600000
HYDROGEN 1.0 3.3185600000 1.4368700000 13.0953100000
HYDROGEN 1.0 4.5549800000 -0.1997300000 11.7165200000
CARBON 6.0 6.1105700000 3.9639000000 22.3866100000
CARBON 6.0 4.1216300000 4.4424400000 21.1020100000
HYDROGEN 1.0 7.8732900000 3.5217100000 21.2520500000
CARBON 6.0 4.7606000000 4.2868500000 22.3363800000
HYDROGEN 1.0 6.6064200000 3.8406000000 23.3428500000
HYDROGEN 1.0 4.2065000000 4.4170700000 23.2667100000
HYDROGEN 1.0 3.0674000000 4.6893500000 21.0889000000
HYDROGEN 1.0 7.4249200000 7.7545300000 18.8583200000
HYDROGEN 1.0 7.6651700000 8.9049700000 17.7652100000
HYDROGEN 1.0 5.3324000000 8.6487800000 17.2222700000
HYDROGEN 1.0 5.5015000000 7.1039000000 18.2759400000
$END
$ECP
MO-ECP GEN 28 3
5 ----- f potential -----
-0.0469492 0 537.9667807
-20.2080084 1 147.8982938
-106.2116302 2 45.7358898
-41.8107368 2 13.2911467
-4.2054103 2 4.7059961
3 ----- s-f potential -----
2.8063717 0 110.2991760
44.5162012 1 23.2014645
82.7785227 2 5.3530131
4 ----- p-f potential -----
4.9420876 0 63.2901397
25.8604976 1 23.3315302
132.4708742 2 24.6759423
57.3149794 2 4.6493040
5 ----- d-f potential -----
3.0054591 0 104.4839977
26.3637851 1 66.2307245
183.3849199 2 39.1283176
98.4453068 2 13.1164437
22.4901377 2 3.6280263
S NONE
S NONE
S NONE
S NONE
S NONE
S NONE
C NONE
C NONE
C NONE
C NONE
C NONE
C NONE
C NONE
C NONE
C NONE
C NONE
H NONE
C NONE
H NONE
C NONE
H NONE
C NONE
H NONE
C NONE
C NONE
C NONE
C NONE
H NONE
H NONE
H NONE
H NONE
C NONE
C NONE
H NONE
C NONE
H NONE
H NONE
H NONE
H NONE
H NONE
H NONE
H NONE
$END
The Input Deck
This is the input deck. It is where you tell GAMESS(US) what job type to execute and where all you individual parameters are entered for your specific job type. The example input deck below is for a geometry optimization and frequency calculation. This input deck is equivalent to the Gaussian job with "opt" and "freq" in the route section.
$CONTRL SCFTYP=RHF RUNTYP=OPTIMIZE DFTTYP=M06-L MAXIT=199 MULT=1 NOSYM=1 ECP=READ $END $SYSTEM TIMLIM=525600 MWORDS=1750 PARALL=.TRUE. $END $BASIS GBASIS=CUSTOMNI EXTFIL=.t. $END $SCF DIRSCF=.TRUE. FDIFF=.f. $END $STATPT OPTTOL=0.0001 NSTEP=500 HSSEND=.t. $END $DATA
An important thing to note is the spacing. In the input deck, there must be 1 space at the beginning of each line of the input deck. If not, the job will fail. Most builders will insert this space anyway, but it help to double check.
The end of the input deck is marked by the "$DATA" line.
Job Title Line
The next line of the file is the job title. It can be anthing you wish, however, we have found that to be on the safe side, we avoide using symbols or spaces
Mo_BDT3
Symmetry Point Group
The next line of the file is the symmetry point group of your molecule. Note that there is no leading space before the point group.
C1
Coordinates
The next block of text is set aside for the coordinates of the molecule. This can be in internal (or z-matrix) format or cartesian coordinates. Note that there is no leading space before the coordinates. One may use the chemical symbol or the full name of each atom in the molecule. Note that the end of the coordinates is signified by an "$END", which MUST have one space preceding it.
MOLYBDENUM 42.0 5.7556500000 4.4039600000 16.5808400000 SULFUR 16.0 7.4169700000 3.1956300000 15.2089300000 SULFUR 16.0 4.0966800000 3.2258300000 15.1761100000 SULFUR 16.0 3.9677300000 4.4940500000 18.3266100000 SULFUR 16.0 7.1776900000 3.5815000000 18.4485200000 SULFUR 16.0 4.3776600000 6.2447400000 15.6786900000 SULFUR 16.0 7.5478700000 6.0679800000 16.2223700000 CARBON 6.0 6.4716900000 2.1004800000 14.1902300000 CARBON 6.0 5.0690300000 2.1781400000 14.1080700000 CARBON 6.0 4.8421800000 4.2701300000 19.8855500000 CARBON 6.0 6.1969000000 3.9249600000 19.9397400000 CARBON 6.0 6.8280600000 3.7834200000 21.1913200000 CARBON 6.0 5.7697600000 7.6933500000 17.4241800000 CARBON 6.0 7.2043100000 7.9413600000 17.8281100000 CARBON 6.0 5.5051400000 7.0409700000 14.5903800000 CARBON 6.0 6.8905200000 6.9194700000 14.7626200000 CARBON 6.0 7.7396400000 7.5379800000 13.8285700000 HYDROGEN 1.0 8.8190700000 7.4520600000 13.9252200000 CARBON 6.0 7.2169400000 8.2960300000 12.7704100000 HYDROGEN 1.0 7.8667000000 8.7825100000 12.0575600000 CARBON 6.0 5.8260300000 8.4502300000 12.6467800000 HYDROGEN 1.0 5.4143000000 9.0544300000 11.8493100000 CARBON 6.0 4.9881500000 7.8192300000 13.5528400000 HYDROGEN 1.0 3.9090500000 7.9420000000 13.4583700000 CARBON 6.0 7.1538500000 1.1569600000 13.4143900000 CARBON 6.0 4.4018100000 1.3603900000 13.1919900000 CARBON 6.0 6.4791600000 0.3185500000 12.5353300000 CARBON 6.0 5.0837400000 0.4369500000 12.4084900000 HYDROGEN 1.0 7.0116000000 -0.4099400000 11.9434600000 HYDROGEN 1.0 8.2399000000 1.0702400000 13.4937600000 HYDROGEN 1.0 3.3185600000 1.4368700000 13.0953100000 HYDROGEN 1.0 4.5549800000 -0.1997300000 11.7165200000 CARBON 6.0 6.1105700000 3.9639000000 22.3866100000 CARBON 6.0 4.1216300000 4.4424400000 21.1020100000 HYDROGEN 1.0 7.8732900000 3.5217100000 21.2520500000 CARBON 6.0 4.7606000000 4.2868500000 22.3363800000 HYDROGEN 1.0 6.6064200000 3.8406000000 23.3428500000 HYDROGEN 1.0 4.2065000000 4.4170700000 23.2667100000 HYDROGEN 1.0 3.0674000000 4.6893500000 21.0889000000 HYDROGEN 1.0 7.4249200000 7.7545300000 18.8583200000 HYDROGEN 1.0 7.6651700000 8.9049700000 17.7652100000 HYDROGEN 1.0 5.3324000000 8.6487800000 17.2222700000 HYDROGEN 1.0 5.5015000000 7.1039000000 18.2759400000 $END
Effective Core Potential Data
The effective core potential (ECP) data is entered after the coordinates. It starts with "$ECP", which must be preceded with a space. The atoms of the molecule are listed in the same order as in the coordinates section and the parameters for the ECP are listed after each atom. Note that for any atom that does NOT have an ECP, one must enter "ECP-NONE" or "NONE" after each atom without an ECP.
$ECP
MO-ECP GEN 28 3
5 ----- f potential -----
-0.0469492 0 537.9667807
-20.2080084 1 147.8982938
-106.2116302 2 45.7358898
-41.8107368 2 13.2911467
-4.2054103 2 4.7059961
3 ----- s-f potential -----
2.8063717 0 110.2991760
44.5162012 1 23.2014645
82.7785227 2 5.3530131
4 ----- p-f potential -----
4.9420876 0 63.2901397
25.8604976 1 23.3315302
132.4708742 2 24.6759423
57.3149794 2 4.6493040
5 ----- d-f potential -----
3.0054591 0 104.4839977
26.3637851 1 66.2307245
183.3849199 2 39.1283176
98.4453068 2 13.1164437
22.4901377 2 3.6280263
S NONE
S NONE
S NONE
S NONE
S NONE
S NONE
C NONE
C NONE
C NONE
C NONE
C NONE
C NONE
C NONE
C NONE
C NONE
C NONE
H NONE
C NONE
H NONE
C NONE
H NONE
C NONE
H NONE
C NONE
C NONE
C NONE
C NONE
H NONE
H NONE
H NONE
H NONE
C NONE
C NONE
H NONE
C NONE
H NONE
H NONE
H NONE
H NONE
H NONE
H NONE
H NONE
$END
Using Effective Core Potentials in GAMESS(US)
For many metal containing compounds, it is very convenient and time saving to use an effective core potential (ECP) for the core metal electrons, as they are usually not important to the reactivity of the complex or the geometry around the metal. Since GAMESS(US) has a limited number of built-in ECPs, one may want to make GAMESS(US) read an external file that contains the ECP data using the "EXTFIL" keyword in the $GBASIS command line of the input file. In addition, to make GAMESS(US) use this external file, one must copy the "rungms" file and modify it accordingly. The following is a list of instructions with commands that will work from a terminal. One could also use WinSCP to do all of this with a GUI rather than a TUI.
Modifiying rungms to Use Custom Basis Set File
1. Copy "rungms" from /scinet/gpc/Applications/gamess to one's own /scratch/$USER/ directory:
cp /scinet/gpc/Applications/gamess/rungms /scratch/$USER/
2. Change to the scratch directory and check to see if "rungms" has copied successfully.
cd /scratch/$USER ls
3. Edit line 147 of the vi.
vi rungms
Move the cursor down to line 147 using the arrow keys. It should say "setenv EXTBAS /dev/null". Using the arrow keys, move the cursor to the first "/" and then hit "i" to insert text. Put the path to your external basis file here. For example, /scratch/$USER/basisset. Then hit "escape". To save the changes and exit vi, type ":" and you should see a colon appear at the bottom of the window. Type "wq" (which should appear at the bottom of the window next to the colon) and then hit enter. Now you are done with vi.
Creating a Custom Basis Set File
1. To create a custom basis set file, you need create a new text document. Our group's common practice is to comment out the first line of this file by inserting an exclamation mark (!) followed by noting the specific basis sets and ECPs that are going to be used for each of the atoms. Let us the molecule Mo(CO)6, Molybdenum hexacarbonyl, as an example. Below is the first line of the the external file, which we will call "CUSTOMMO" (NOTE: you can use any name for the external file that suits you, as long as it has no spaces and is 8 characters or less).
! 6-31G on C and O and LANL2D2 ECP on Mo
2. The next step is to visit the EMSL Basis Set exchange and select C and O from the periodic table. Then, on the left of the page, select "6-31G" as the basis set. Finally, make sure the output is in GAMESS(US) format using the drop-down menu and then click "get basis set".
3. A new window should appear with text in it. For our example case, the text looks like this:
! 6-31G EMSL Basis Set Exchange Library 10/13/09 11:12 AM ! Elements References ! -------- ---------- ! H - He: W.J. Hehre, R. Ditchfield and J.A. Pople, J. Chem. Phys. 56, ! Li - Ne: 2257 (1972). Note: Li and B come from J.D. Dill and J.A. ! Pople, J. Chem. Phys. 62, 2921 (1975). ! Na - Ar: M.M. Francl, W.J. Petro, W.J. Hehre, J.S. Binkley, M.S. Gordon, ! D.J. DeFrees and J.A. Pople, J. Chem. Phys. 77, 3654 (1982) ! K - Zn: V. Rassolov, J.A. Pople, M. Ratner and T.L. Windus, J. Chem. Phys. ! 109, 1223 (1998) ! Note: He and Ne are unpublished basis sets taken from the Gaussian ! program ! $DATA
CARBON S 6 1 3047.5249000 0.0018347 2 457.3695100 0.0140373 3 103.9486900 0.0688426 4 29.2101550 0.2321844 5 9.2866630 0.4679413 6 3.1639270 0.3623120 L 3 1 7.8682724 -0.1193324 0.0689991 2 1.8812885 -0.1608542 0.3164240 3 0.5442493 1.1434564 0.7443083 L 1 1 0.1687144 1.0000000 1.0000000
OXYGEN S 6 1 5484.6717000 0.0018311 2 825.2349500 0.0139501 3 188.0469600 0.0684451 4 52.9645000 0.2327143 5 16.8975700 0.4701930 6 5.7996353 0.3585209 L 3 1 15.5396160 -0.1107775 0.0708743 2 3.5999336 -0.1480263 0.3397528 3 1.0137618 1.1307670 0.7271586 L 1 1 0.2700058 1.0000000 1.0000000 $END
3. Now, copy and paste the text between the $DATA and $END headings onto our external text file, CUSTOMMO. We also need to change the change the name of each element to it symbol in the periodic table. Finally, we need to add the name of the external file next to the element symbol, separated by one space. Note that there should be a blank line separating the basis set information and the first, commented-out line (The line starting with the '!'). The CUSTOMMO should look like this:
! 6-31G on C and O and LANL2D2 ECP on Mo
C CUSTOMMO S 6 1 3047.5249000 0.0018347 2 457.3695100 0.0140373 3 103.9486900 0.0688426 4 29.2101550 0.2321844 5 9.2866630 0.4679413 6 3.1639270 0.3623120 L 3 1 7.8682724 -0.1193324 0.0689991 2 1.8812885 -0.1608542 0.3164240 3 0.5442493 1.1434564 0.7443083 L 1 1 0.1687144 1.0000000 1.0000000
O CUSTOMMO S 6 1 5484.6717000 0.0018311 2 825.2349500 0.0139501 3 188.0469600 0.0684451 4 52.9645000 0.2327143 5 16.8975700 0.4701930 6 5.7996353 0.3585209 L 3 1 15.5396160 -0.1107775 0.0708743 2 3.5999336 -0.1480263 0.3397528 3 1.0137618 1.1307670 0.7271586 L 1 1 0.2700058 1.0000000 1.0000000
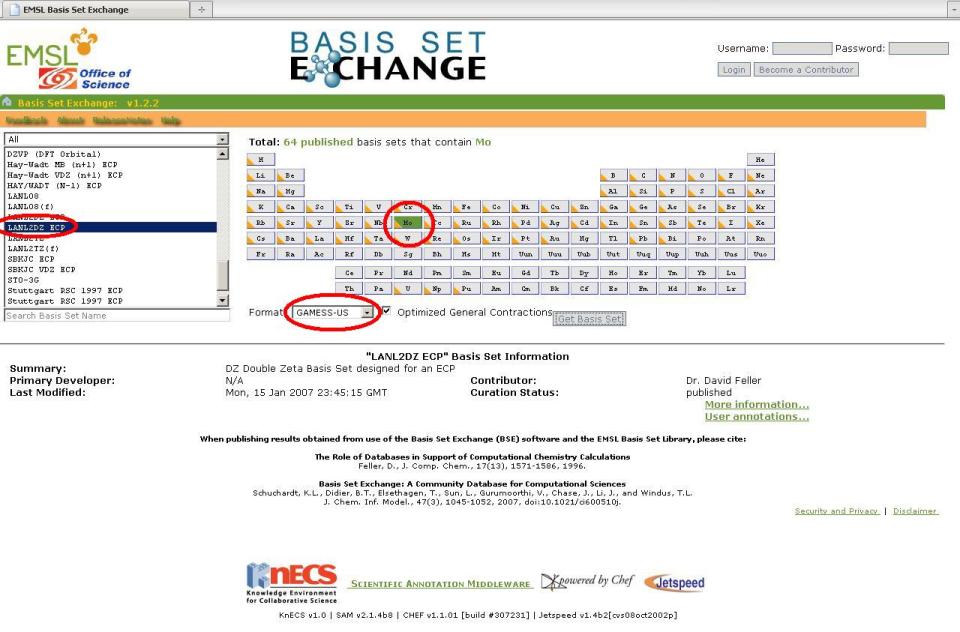
4. Repeat Step 3 above but choose Mo and select the LANL2DZ ECP instead. A new window will pop up with the basis set information as well as the ECP data we need, since we specified the LANL2DZ ECP. The ECP data is not inserted into the external file, rather it is placed into the input file itself (More on this later).
5. After copying the molybdenum basis set information, your fiished external basis set file should look like this:
! 6-31G on C and O and LANL2D2 ECP on Mo
C CUSTOMMO S 6 1 3047.5249000 0.0018347 2 457.3695100 0.0140373 3 103.9486900 0.0688426 4 29.2101550 0.2321844 5 9.2866630 0.4679413 6 3.1639270 0.3623120 L 3 1 7.8682724 -0.1193324 0.0689991 2 1.8812885 -0.1608542 0.3164240 3 0.5442493 1.1434564 0.7443083 L 1 1 0.1687144 1.0000000 1.0000000
O CUSTOMMO S 6 1 5484.6717000 0.0018311 2 825.2349500 0.0139501 3 188.0469600 0.0684451 4 52.9645000 0.2327143 5 16.8975700 0.4701930 6 5.7996353 0.3585209 L 3 1 15.5396160 -0.1107775 0.0708743 2 3.5999336 -0.1480263 0.3397528 3 1.0137618 1.1307670 0.7271586 L 1 1 0.2700058 1.0000000 1.0000000
Mo CUSTOMO S 3 1 2.3610000 -0.9121760 2 1.3090000 1.1477453 3 0.4500000 0.6097109 S 4 1 2.3610000 0.8139259 2 1.3090000 -1.1360084 3 0.4500000 -1.1611592 4 0.1681000 1.0064786 S 1 1 0.0423000 1.0000000 P 3 1 4.8950000 -0.0908258 2 1.0440000 0.7042899 3 0.3877000 0.3973179 P 2 1 0.4995000 -0.1081945 2 0.0780000 1.0368093 P 1 1 0.0247000 1.0000000 D 3 1 2.9930000 0.0527063 2 1.0630000 0.5003907 3 0.3721000 0.5794024 D 1 1 0.1178000 1.0000000
-- mzd 13 October 2009
Climate Modelling
Medicine/Bio
High Energy Physics
Structural Biology
Molecular simulation of proteins, lipids, carbohydrates, and other biologically relevant molecules.
Molecular Dynamics (MD) simulation
GROMACS
Please refer to the GROMACS page
NAMD
NAMD is one of the better scaling MD packages out there. With sufficiently large systems, it is able to scale to hundreds or thousands of cores on Scinet. Below are details for compiling and running NAMD on Scinet.
More information regarding performance and different compile options coming soon...
Compiling NAMD for GPC
Ensure the proper compiler/mpi modules are loaded. <source lang="sh"> module load intel module load openmpi/1.3.3-intel-v11.0-ofed </source>
Compile Charm++ and NAMD <source lang="sh">
- Unpack source files and get required support libraries
tar -xzf NAMD_2.7b1_Source.tar.gz cd NAMD_2.7b1_Source tar -xf charm-6.1.tar wget http://www.ks.uiuc.edu/Research/namd/libraries/fftw-linux-x86_64.tar.gz wget http://www.ks.uiuc.edu/Research/namd/libraries/tcl-linux-x86_64.tar.gz tar -xzf fftw-linux-x86_64.tar.gz; mv linux-x86_64 fftw tar -xzf tcl-linux-x86_64.tar.gz; mv linux-x86_64 tcl
- Compile Charm++
cd charm-6.1 ./build charm++ mpi-linux-x86_64 icc --basedir /scinet/gpc/mpi/openmpi/1.3.3-intel-v11.0-ofed/ --no-shared -O -DCMK_OPTIMIZE=1 cd ..
- Compile NAMD.
- Edit arch/Linux-x86_64-icc.arch and add "-lmpi" to the end of the CXXOPTS and COPTS line.
- Make a builds directory if you want different versions of NAMD compiled at the same time.
mkdir builds ./config builds/Linux-x86_64-icc --charm-arch mpi-linux-x86_64-icc cd builds/Linux-x86_64-icc/ make -j4 namd2 # Adjust value of j as desired to specify number of simultaneous make targets. </source> --Cmadill 16:18, 27 August 2009 (UTC)
Running Fortran
On the development nodes, there is an old gcc. The associated libraries are not on the compute nodes. Ensure the line:
module load gcc
is in your .bashrc file.