BGQ
| Blue Gene/Q (BGQ) | |
|---|---|
| Installed | August 2012 |
| Operating System | RH6.2, CNK (Linux) |
| Number of Nodes | 2048(32,768 cores), 512 (8,192 cores) |
| Interconnect | 5D Torus (jobs), QDR Infiniband (I/O) |
| Ram/Node | 16 GB |
| Cores/Node | 16 (64 threads) |
| Login/Devel Node | bgqdev-fen1,bgq-fen1 |
| Vendor Compilers | bgxlc, bgxlf |
| Queue Submission | Loadleveler |
System Status: BETAOct 25, 2012: The development system driver will be upgraded to R1V1M2 on Friday, and will be unavailable for about 2 hours. The production system will still be accessible (from login.scinet.utoronto.ca). POSTPONED TILL NEXT WEEK. Oct 24, 2012: The production system driver has been upgraded to R1V1M2 and is available again. The development system is still at R1V1M1. Oct 23, 2012: The production system is being upgraded and unavailable. The development system remains up. Oct 12, 2012: Updated wiki. Now in friendly-user mode! Oct 3, 2012: The BGQ is in Beta testing and as such the environment is still under development as is the wiki information contained here. |
Support Email
Please use bgq-support@scinethpc.ca for BGQ specific inquiries.
Specifications
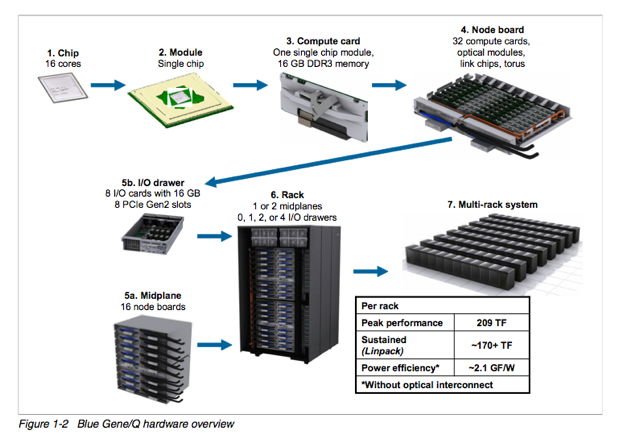
BGQ is an extremely dense and energy efficient 3rd generation blue gene IBM supercomputer built around a system on a chip compute node that has a 16core 1.6GHz PowerPC based CPU (PowerPC A2) with 16GB of Ram. The nodes are bundled in groups of 32 into a node board (512 cores), and 16 boards make up a midplane (8192 cores) with 2 midplanes per rack, or 16,348 cores and 16 GB of RAM per rack. The compute nodes run a very lightweight linux based operating system called CNK. The compute nodes are all connected together using a custom 5D torus highspeed interconnect. Each rack has 16 I/O nodes that run a full Redhat Linux OS that manages the compute nodes and mounts the filesystem. SciNet has 2 BGQ systems, a half rack 8192 core development system, and a 2 rack 32,768 core production system.
5D Torus Network
The network topology of Blue/Gene Q is a five-dimensional (5D) torus, with direct links between the nearest neighbors in the ±A, ±B, ±C, ±D, and ±E directions. As such there are only a few optimum block sizes that will use the network efficiently.
| Node Boards | Compute Nodes | Cores | Torus Dimensions |
| 1 | 32 | 512 | 2x2x2x2x2 |
| 2 (adjacent pairs) | 64 | 1024 | 2x2x4x2x2 |
| 4 (quadrants) | 128 | 2048 | 2x2x4x4x2 |
| 8 (halves) | 256 | 4096 | 4x2x4x4x2 |
| 16 (midplane) | 512 | 8192 | 4x4x4x4x2 |
| 32 (1 rack) | 1024 | 16384 | 4x4x4x8x2 |
| 64 (2 racks) | 2048 | 32768 | 4x4x8x8x2 |
Login/Devel Nodes
The development nodes for the BGQ are bgqdev-fen1 for the half-rack development system and bgq-fen1 for the 2-rack production system. You can login to them from the regular login.scinet.utoronto.ca login nodes or directly from outside to the half-rack development system using bgqdev.scinet.utoronto.ca.
These development nodes are Power7 running Linux which serve as compilation and submission hosts for the BGQ. Programs are cross-compiled for the BGQ on these nodes and then submitted to the queue using loadleveler.
Modules and Environment Variables
To use most packages on the SciNet machines - including most of the compilers - , you will have to use the `modules' command. The command module load some-package will set your environment variables (PATH, LD_LIBRARY_PATH, etc) to include the default version of that package. module load some-package/specific-version will load a specific version of that package. This makes it very easy for different users to use different versions of compilers, MPI versions, libraries etc.
A list of the installed software can be seen on the system by typing
$ module avail
To load a module (for example, the default version of the intel compilers)
$ module load vacpp
To unload a module
$ module unload vacpp
To unload all modules
$ module purge
These commands can go in your .bashrc files to make sure you are using the correct packages.
Note that a module load command only sets the environment variables in your current shell (and any subprocesses that the shell launches). It does not effect other shell environments.
If you always require the same modules, it is easiest to load those modules in your .bashrc and then they will always be present in your environment; if you routinely have to flip back and forth between modules, it is easiest to have almost no modules loaded in your .bashrc and simply load them as you need them (and have the required module load commands in your job submission scripts).
Compilers
The BGQ uses IBM XL compilers to cross-compile code for the BGQ. Compilers are available for FORTRAN, C, and C++. They are accessible by default, or by loading the xlf and vacpp modules. The compilers by default produce static binaries, however with BGQ it is possible to now use dynamic libraries as well. The compilers follow the XL conventions with the prefix bg, so bgxlc and bgxlf90 are the C and FORTRAN compilers respectively.
Most users however will use the MPI variants, i.e. mpixlf90 and mpixlc and which are available by loading the mpich2 module.
module load mpich2
It is recommended to use at least the following flags when compiling and linking
-O3 -qarch=qp -qtune=qp
Software modules installed on the BGQ
| Software | Version | Comments | Command/Library | Module Name |
|---|---|---|---|---|
| Compilers | ||||
| IBM fortran compiler | 14.1 | These are cross compilers | bgxlf,bgxlf_r,bgxlf90,... | xlf |
| IBM c/c++ compilers | 12.1 | These are cross compilers | bgxlc,bgxlC,bgxlc_r,bgxlC_r,... | vacpp |
| MPICH2 MPI library | 1.4.1 | There are 4 versions (see BGQ Applications Development document). | mpicc,mpicxx,mpif77,mpif90 | mpich2 |
| GCC Compiler | 4.4.6 | GNU Compiler Collection for BGQ | powerpc64-bgq-linux-gcc, powerpc64-bgq-linux-g++, powerpc64-bgq-linux-gfortran | bgqgcc |
| Debug/performance tools | ||||
| gdb | 7.1 | GNU Debugger | gdb | gdb |
| Storage tools/libraries | ||||
| Libraries | ||||
| ESSL | 5.1 | IBM Engineering and Scientific Subroutine Library (manual below) | libesslbg,libesslsmpbg | essl |
| FFTW | 2.1.5, 3.3.2, 3.1.2-esslwrapper | Fast fourier transform | libsfftw,libdfftw,libfftw3, libfftw3f | fftw/2.1.5, fftw/3.3.2, fftw/3.1.2-esslwrapper |
| GSL | 1.15 | GNU Scientific Library | libgsl, libgslcblas | gsl |
Job Submission
As the BGQ architecture is different from the development nodes, the only way to test your program is to submit a job to the BGQ. Jobs are submitted through loadleveler using runjob which in many ways similar to mpirun or mpiexec. As shown above in the network topology overview, there are only a few optimum job size configurations which is also further constrained by each block requiring a minimum of one IO node. In SciNet's configuration (with 8 I/O nodes per midplane) this allows 64 nodes (1024 cores) to be the smallest block size. Normally a block size matches the job size to offer fully dedicated resources to the job. Smaller jobs can be run within the same block however this results in shared resources (network and IO) and are referred to as sub-block jobs and are described in more detail below.
runjob
All BGQ jobs are launced using runjob which for those familiar with MPI is analogous to mpirun/mpiexec. The "block" argument is the predefined group of nodes that have already been configured and booted and when using loadleveler this is set for you. For this example block R00-M0-N03-64 is made up of 2 node cards with 64 compute nodes (1024 cores). To run a job with 16 tasks/node and 1024 total tasks the following command is used.
runjob --block BLOCK_ID --ranks-per-node=16 --np 1024 --cwd=$PWD : $PWD/code -f file.in
For pure mpi jobs, it is advisable always to give the number of ranks per node, because the default value of 1 may leave 15 cores on the node idle. the argument to ranks-per-node may be 1, 2, 4, 8, 16, 32, or 64.
runjob flags are shown with
runjob -h
a particularly useful one is
--verbose #
where # is from 1-7 which can be helpful in debugging an application.
Batch Jobs
Job submission is done through loadleveler with a few blue gene specific commands. The command "bg_size" is in number of nodes, not cores, so a bg_size=64 would be 1024 cores.
#!/bin/sh # @ job_name = bgsample # @ job_type = bluegene # @ comment = "BGQ Job By Size" # @ error = $(job_name).$(Host).$(jobid).err # @ output = $(job_name).$(Host).$(jobid).out # @ bg_size = 64 # @ wall_clock_limit = 30:00 # @ bg_connectivity = Torus # @ queue # Launch all BGQ jobs using runjob runjob --np 1024 --ranks-per-node=16 --envs OMP_NUM_THREADS=1 --cwd=$SCRATCH/ : $HOME/mycode.exe myflags
To submit to the queue use
llsubmit myscript.sh
to see running jobs
llq -b
to cancel a job use
llcancel JOBID
and to look at details of the bluegene resources use
llbgstatus
Note: the loadleveler script commands are not run on a bgq compute node but on the front-end node. Only programs started with runjob run on the bgq compute nodes. You should therefore keep scripting in the submission script to a bare minimum.
Interactive Use
As BGQ codes are cross-compiled they cannot be run direclty on the front-nodes. Users however only have access to the BGQ through loadleveler which is appropriate for batchjobs, however an interactive session is typically beneficial when debugging and developing. As such a script has been written to allow a session in which runjob can be run interactively. The script uses loadleveler to setup a block and set all the correct environment variables and then launch a spawned shell on the front-end node. The debugjob session currently allows a 30 minute session on 64 nodes.
[user@bgqdev-fen1]$ debugjob [user@bgqdev-fen1]$ runjob --np 64 --ranks-per-node=16 --cwd=$PWD : $PWD/my_code -f myflags [user@bgqdev-fen1]$ exit
NOTE: This is a prototype script so be gentle
Sub-block jobs
BGQ allows sub-block jobs, however this would need to be done from within the same loadleveler submission scripts using multiple calls to runjob. To run a sub-block job (ie share a block) you need to specify a "--corner" within the block to start the job and a 5D AxBxCxDxE "--shape". The following example shows 2 jobs sharing the same block.
runjob --block R00-M0-N03-64 --corner R00-M0-N03-J00 --shape 1x1x1x2x2 --ranks-per-node=16 --np 64 --cwd=$PWD : $PWD/code -f file.in runjob --block R00-M0-N03-64 --corner R00-M0-N03-J04 --shape 2x2x2x2x1 --ranks-per-node=16 --np 256 --cwd=$PWD : $PWD/code -f file.in
Filesystem
The BGQ has its own dedicated 500TB GPFS filesystem for both $HOME and $SCRATCH. Currently, quotas are set to 50GB and 20TB and neither of these is backed up (the plan is to have a backup system for /home eventually).
While this file system is shared between the bgq development and production system, the file system is not shared with the other SciNet systems (gpc, tcs, p7, arc), nor is the other file system mounted on the BGQ. Use scp to copy files from one file system to the other, e.g., from bgqdev-fen1, you could do
$ scp @login.scinet.utoronto.ca:code.tgz .
or from a login node you could do
$ scp code.tgz @bgqdev.scinet.utoronto.ca:
Note that you currently have to use the full names of these systems, and that it will ask for your password.
Documentation
- Julich Documentation
- BGQ System Administration Guide
- BGQ Application Development
- IBM XL C/C++ for Blue Gene/Q: Getting started
- IBM XL C/C++ for Blue Gene/Q: Compiler reference
- IBM XL C/C++ for Blue Gene/Q: Language reference
- IBM XL C/C++ for Blue Gene/Q: Optimization and Programming Guide
- IBM XL Fortran for Blue Gene/Q: Getting started
- IBM XL Fortran for Blue Gene/Q: Compiler reference
- IBM XL Fortran for Blue Gene/Q: Language reference
- IBM XL Fortran for Blue Gene/Q: Optimization and Programming Guide
- IBM ESSL (Engineering and Scientific Subroutine Library) 5.1 for Linux on Power